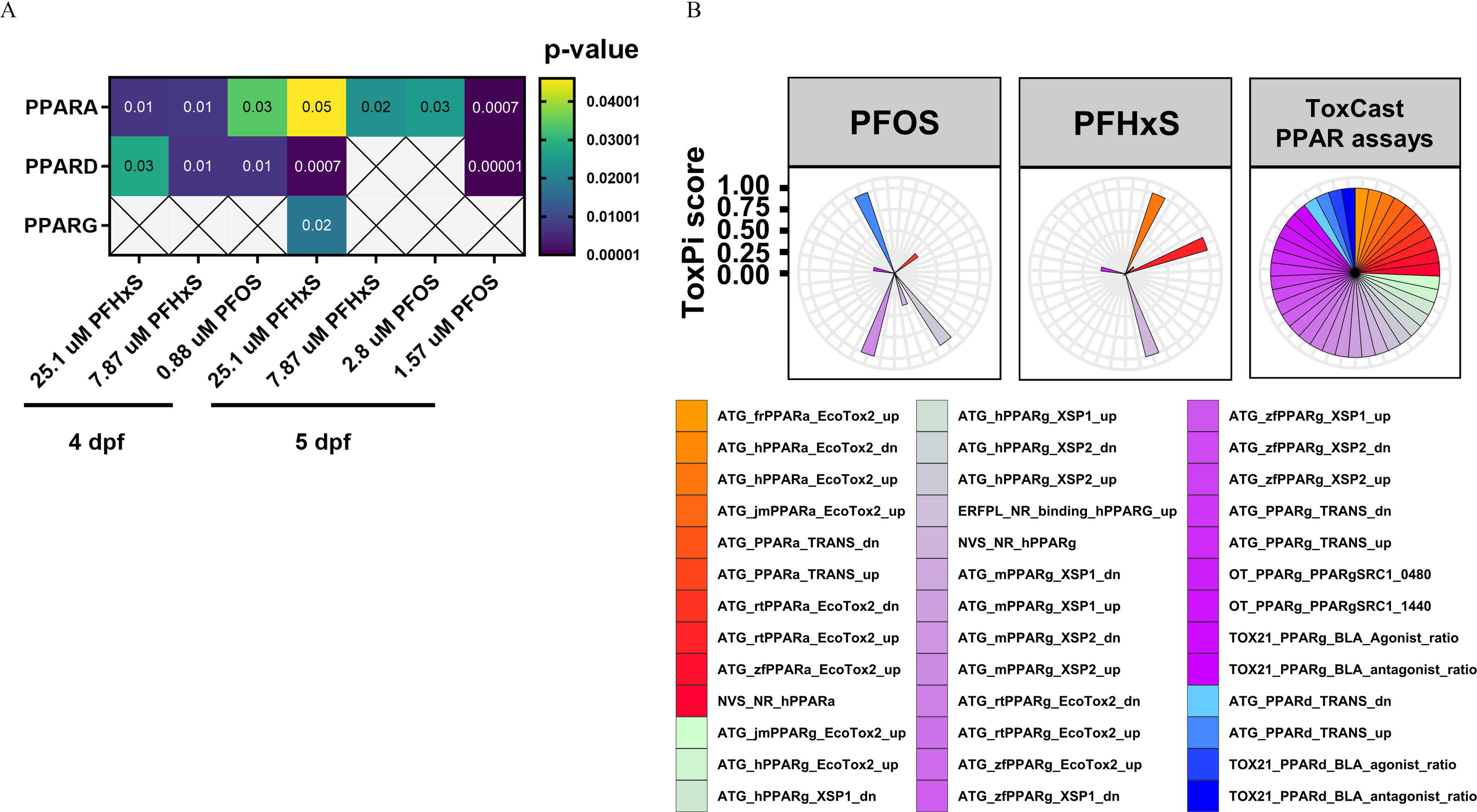
Fig. 6 Predicted upstream regulators affected by developmental exposure to PFOS or PFHxS and in vitro potency profiles. (A) Human orthologs for zebrafish differentially expressed genes (DEGs) were analyzed by Ingenuity Pathway Analysis and predicted human PPARα, PPARδ, and PPARγ as putative upstream regulators for multiple concentrations of PFOS and PFHxS at 4–5 dpf. Significant enrichment was identified by Fisher’s exact test 𝑝<0.05; crosses indicate nonsignificance. Five biological replicates containing pools of 15 heads were used. (B) Visualized potency profiles of PFOS and PFHxS in US EPA CompTox (version 3.5) in vitro assays targeting human (h), rat (r), mouse (m), zebrafish (zf), or Japanese medaka (jm) PPARα, PPARδ, and/or PPARγ. Profiles for both chemicals were calculated based on AC50-values using the ToxPi framework. Distance from the center indicates potency. All assays were weighted equally. ToxPi data can be found in Excel Table S19. Note: AC50, 50% activity concentration; dpf, days postfertilization; EPA, Environmental Protection Agency; PFHxS, perfluorohexanesulfonic acid; PFOS, perfluorooctanesulfonic acid; PPAR, peroxisome proliferator-activated receptor; ToxPi, toxicity prioritization index.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Environ. Health Perspect.