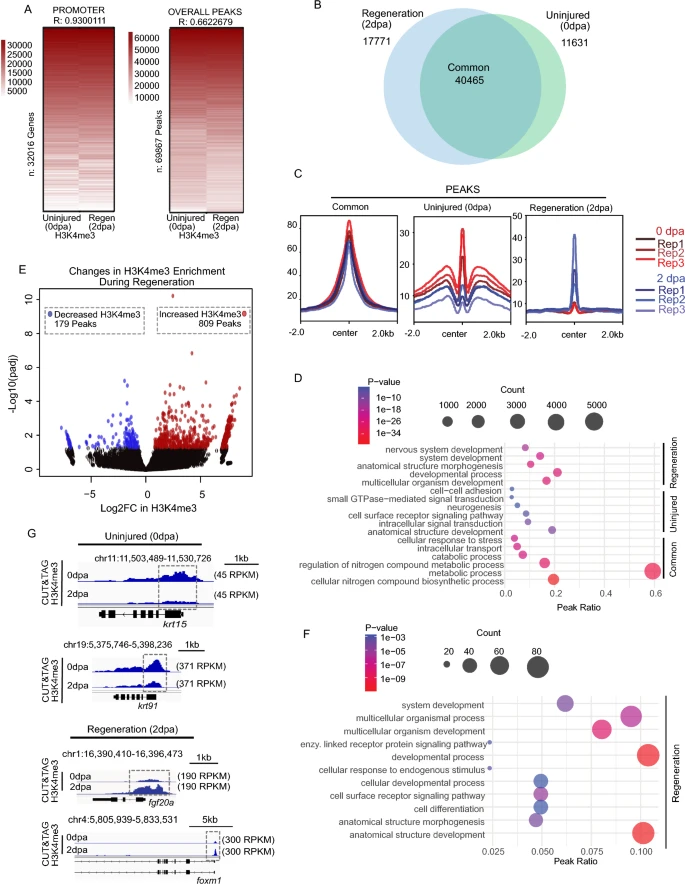
Fig. 3 CUT&Tag detects H3K4me3 at the TSS of annotated genes in regenerating (2 dpa) amputated zebrafish fin. a Heatmap of Pearson correlation values demonstrating significant correlation between uninjured (0 dpa) and regenerating fins (2 dpa) both within promoters and at peak regions. b Venn diagram depicting shared and unique H3K4me3 peaks between Uninjured (0 dpa), and Regeneration (2 dpa) zebrafish caudal fins samples. c Profile plots of H3K4me3 and chromatin accessibility enrichment at classified peak regions (Common, Uninjured (0 dpa), and Regeneration (2 dpa)). d Gene ontology analysis of classified H3K4me3 marked regions, including Common, Uninjured (0 dpa), and Regeneration (2 dpa) specific regions based on Venn diagram. e Volcano plot of H3K4me3 enrichment changes comparing uninjured and regenerating fin (significance thresholds = absolute Log2FC > 0.5 and adjusted p-value < 0.05). f Gene ontology analysis of H3K4me3 changes in 2 dpa-specific region with at least 0.05 FDR based on DESeq2 result. g Genome browser view showing enrichment of H3K4me3 at putative regulatory elements for selected genes in Uninjured (0 dpa), and Regeneration (2 dpa) classes
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Epigenetics Chromatin