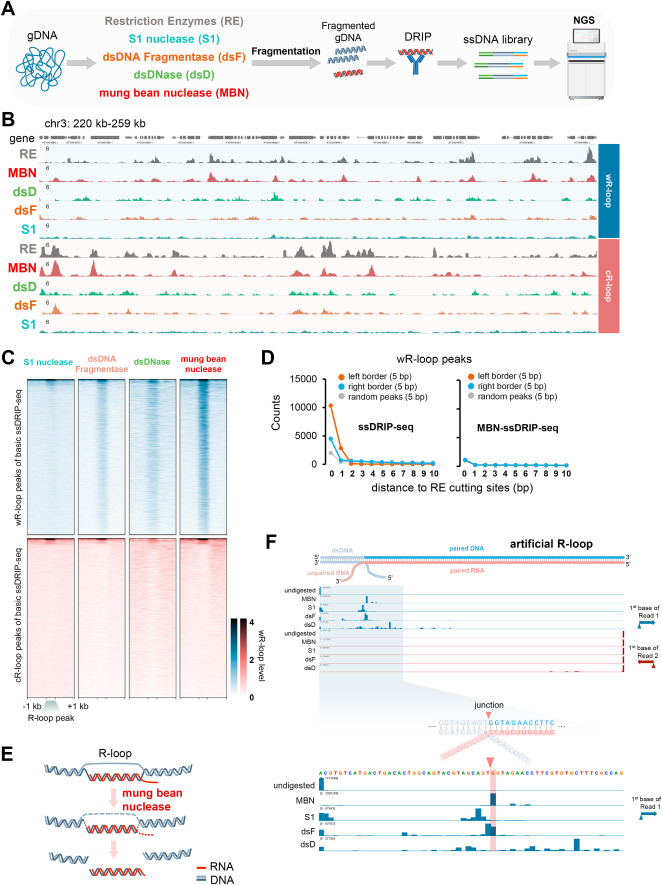
Fig. 1 Improvements of ssDRIP-seq by optimizing the genomic DNA fragmentation. A. Schematic of the exploration experiment for the improvement of ssDRIP-seq. Briefly, the gDNA digestion step was first performed by using S1 nuclease, dsDNA Fragmentase, dsDNase, mung bean nuclease, and restriction enzymes. Then gDNA was fragmented by sonication, followed by DRIP, ssDNA library preparation, and sequencing. B. IGV snapshot of Arabidopsis wR-loop (Watson R-loop, with a Watson strand DNA as the unpaired ssDNA) and cR-loop (Crick R-loop, with a Crick strand DNA as the unpaired ssDNA) signals from basic ssDRIP-seq using restriction enzymes (RE, grey) and other modified ssDRIP-seq using mung bean nuclease (MBN, red), dsDNase (dsD, light green), dsDNA Fragmentase (dsF, brown), or S1 nuclease (S1, cyan) for gDNA treatment. C. Heatmaps of Arabidopsis wR-loop signals of modified ssDRIP-seq using S1 nuclease, dsDNA Fragmentase, dsDNase, or mung bean nuclease. The signals around wR-loop (upper, blue) or cR-loop (lower, red) peaks of basic ssDRIP-seq were shown. D. Statistics of boundaries of wR-loop peaks (extended ± 5 bp) from basic ssDRIP-seq (left) or MBN-ssDRIP-seq (right, modified ssDRIP-seq using mung bean nuclease) with different distances from restriction enzyme cutting sites (DdeI, MseI, NlaIII and MboI). 0 bp means overlapping. E. Supposed model of R-loop treatment by mung bean nuclease. F. MBN-ssDRIP-seq result of artificial half R-loop (seen in Fig. S1H). The artificial R-loop was first treated with mung bean nuclease (MBN), S1 nuclease(S1), dsDNA Fragmentase (dsF), or dsDNase (dsD), followed by DRIP and ssDNA library preparation. IGV snapshot showed the counts of the first bases of Watson (blue) and Crick (red) reads. The junction and expected boundaries was marked by red triangle and red shadow respectively.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Insight