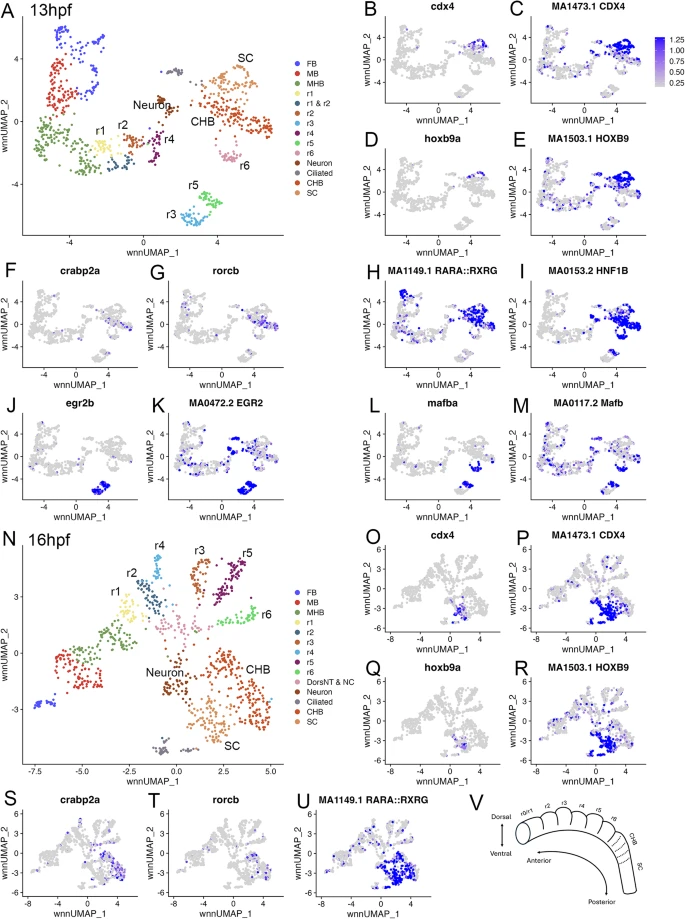
Fig. 1 The caudal hindbrain is molecularly resolved at 13hpf and 16hpf in zebrafish. See also Additional file 1, Table S1. (A) UMAP of 13hpf neural clusters. (B-M.) Feature plots showing expressing of the indicated genes (B, D, F, G, J, L), or chromVar activity of the indicated TF motifs (C, E, H, I, K, M). (N) UMAP of 16hpf neural clusters. (O-U.) Feature plots showing expressing of the indicated genes (O, Q, S, T), or chromVar activity of the indicated TF motifs (P, R, U). (V) Schematic of zebrafish hindbrain with embryonic axes indicated. Dashed lines indicate boundaries delineating previously postulated pseudo-rhombomeres. UMAPs in A, N are based on 8.0 and 6.0 res clustering, respectively (chosen because this fully resolves r1-r6). In cases where multiple clusters were assigned the same identity, as was the case for FB, MB, MHB, CHB, and SC, they were combined into a single cluster labeled with that identity. Abbreviations used in all UMAPs: CHB - caudal hindbrain, HB - hindbrain, FB - forebrain, MB - midbrain, MHB - midbrain-hindbrain boundary, NT - neural tube, NC - neural crest, r - rhombomere, SC - spinal cord
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Neural Dev.