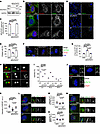
Fig. 1 Loss of NDUFAF2 in RPE cells results in primary cilia defects. (A) NDUFAF2WT was stably expressed in NDUFAF2–/– cells. Western blot analysis was performed with antibodies against NDUFAF2 and GAPDH. (B) Cells stained with TOMM20 (green) and NDUFAF2 (red) antibodies. DNA stained with DAPI (blue). Scale bar: 10 μm. (C) NDUFAF2 staining of mitochondria in wild-type, NDUFAF2–/–, and NDUFAF2WT-re-expressing RPE1 cells. (D) Cells stained with polyglutamylated tubulin (green) antibodies. DNA stained with DAPI (blue). Scale bar: 10 μm. (E) Percentage of ciliated cells after serum starving for 2 days; >150 cells analyzed for each independent experiment. (F) Immunofluorescent analysis of cells serum-starved for 2 days. Cells stained with CP110 (red), CEP164 (green), and FOP (FGFR1 oncogene partner; blue) antibodies. DNA stained with DAPI (blue). Scale bars: 10 μm. (G) Graph shows percentage of serum-starved cells with two CP110 dots at the centrioles; >150 cells analyzed for each independent experiment. (H) Immunofluorescent analysis of cells serum-starved for 6 hours. Cells were stained with myosin-Va (red) and centrin (green) antibodies. DNA stained with DAPI (blue). Scale bar: 2 μm. (I) Percentage of cells with ciliary vesicles demonstrated by an antibody against myosin-Va after serum starving for 6 hours (CV, ciliary vesicle; NCV, no ciliary vesicle; PCV, preciliary vesicle); >150 cells analyzed for each independent experiment. (J) Immunostaining of cells serum-starved for 2 days. Scale bars: 10 μm. (K) Quantification of NPHP1 signal intensity at the centrioles; >100 cells analyzed for each independent experiment. (L) Immunofluorescent analysis of cells serum-starved for 2 days. Cells stained with TCTN2 (red) and pGlu-Tu (green) antibodies. DNA stained with DAPI (blue). Scale bars: 10 μm, 2 μm. (M) Quantification of TCTN2 signal intensity at the centrioles; >50 cells analyzed for each independent experiment. (N) Immunofluorescent analysis of cells serum-starved for 2 days. Cells stained with MKS1 (red) and pGlu-Tu (green) antibodies. DNA stained with DAPI (blue). Scale bars: 10 μm, 2 μm. (O) Quantification of MKS1 signal intensity at the centrioles; >50 cells analyzed for each independent experiment. Bars represent mean ± SD, n = 3. Exact P values are indicated. ANOVA followed by Tukey-Kramer multiple-comparison test.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Journal of Clin. Invest.