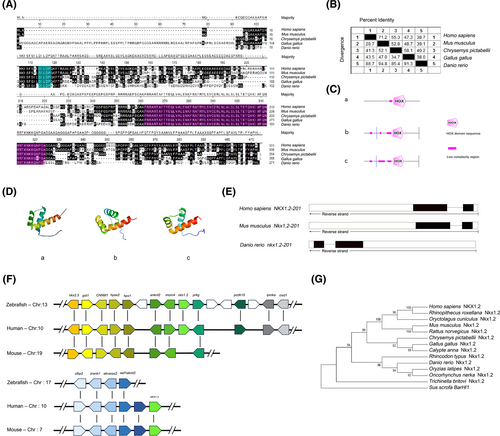
Fig. 1 Nkx1-2 proteins have a highly conserved homeodomain. (A) Alignment of the Nkx1-2 protein sequence. Black color letters indicate the same amino acids. The homeodomain sequences were purple, whereas the -DILDP- motif sequences were blue. (B) Similarity analysis of Nkx1-2 amino acid sequences of different species. (C) SMART program was used to analyze the secondary structure of Nkx1-2 proteins. (D) SWISS-MODEL website (http://www.expasy.org/) analyzes the three-dimensional structure of the zebrafish, mice, and human Nkx1-2 proteins. (E) Comparison of gene organization of Nkx1-2 genes in different species. Boxes represent exons and horizontal lines represent introns. The coding exons were marked in black boxes, whereas the noncoding exons were white boxes. (F) Syntenic map of the Nkx1-2 genes in the chromosomes. Boxes represent genes, and transcription direction is indicated by arrow. (G) Evolutionary analysis of Nkx1-2. A phylogenetic tree was constructed using the neighbor-joining method of the MEGA 6.0 software. Each node was calculated 1000 times, and the number on the node represents the calculated percentage value. Sus scrofa BarHl1 used as the outgroup. See Table S1 for the sequence source. [Color figure can be viewed at wileyonlinelibrary.com]
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Obesity (Silver Spring)