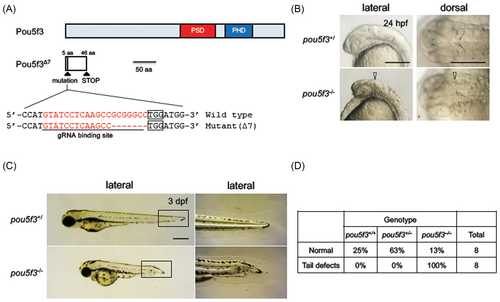
Fig. 1 Disruption and activation of the functions of Pou5f3 in zebrafish. (A) The schematic view of the wild-type and mutant Pou5f3 (Pou5f3∆7). The coding region and the nonsense region due to a frame shift are shown with light blue and white boxes, respectively. The target site of CRISPR/Cas9-mediated disruption is shown below the scheme of the Pou5f3 protein. The sense sequence for the gRNA and PAM sequences are shown with an underline and boxes, respectively. The disruption was found to be a 7-bp deletion that will cause a frameshift mutation (pou5f3∆7). PSD, POU-specific domain; PHD, POU homeodomain. (B) Phenotypes observed in the heads of pou5f3 mutants at 24 hpf. Disruption of the isthmus is shown with triangles. (C) Phenotypes observed in pou5f3 mutants at 3 dpf. The boxed areas in the left panels are enlarged on the right. (D) Morphology of the tail at 3 dpf for different genotypes (wild-type, heterozygotes, and homozygotes). Eight of normal embryos and tail-deformed embryos were randomly selected and genotyped by HMA. HMA, heteroduplex mobility assay.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Dev. Dyn.