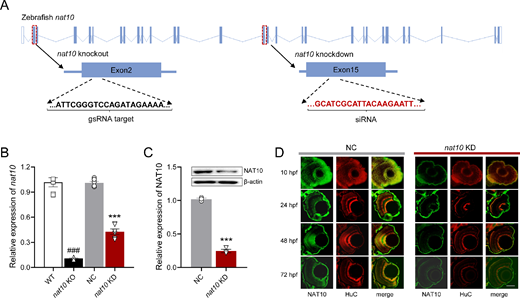
Fig. 3 Disruption of nat10 in zebrafish via CRISPR/Cas9 and siRNA. (A) Illustrates the genomic structure of the zebrafish nat10 gene, highlighting the CRISPR/Cas9 target site in exon 2 (orange underline) and the siRNA target site in exon 15 (red underline) for knockout and knockdown, respectively. (B) Quantitative analysis of nat10 mRNA expression levels in wild-type (WT), nat10 knockout (KO), negative control (NC), and nat10 knockdown (KD) zebrafish using real-time PCR. Results are presented as mean ± SEM from six independent experiments. Statistical analysis was performed using 1-way ANOVA with the Tukey post hoc test; ###P < 0.001 vs. WT, ***P < 0.001 vs. NC. (C) Western blot analysis of NAT10 protein expression in NC and nat10 KD zebrafish. Data are expressed as mean ± SEM from 3 independent experiments, analyzed with an unpaired two-tailed Student’s t-test; **P < 0.01 vs. NC. (D) Histological sections stained with NAT10 (green) and HuC (red) from NC and nat10 KD zebrafish retinas at 10 hpf, 24 hpf, 48 hpf, and 72 hpf. Scale bar = 50 µm.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Invest. Ophthalmol. Vis. Sci.