Figure Caption
Figure 6
Snf8 loss of function in zebrafish leads to developmental defects that are rescued by expression of the wild-type SNF8 cDNA but not disease-associated variants, and causes reduced anterior brain area and ectopic proliferation
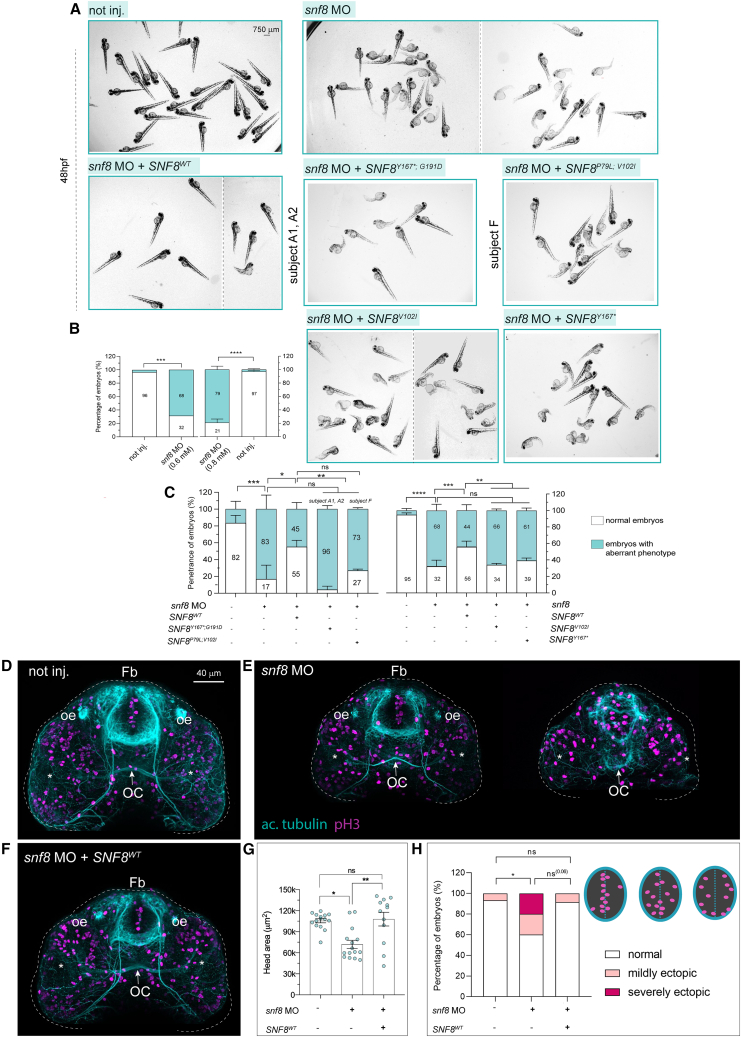
(A) Representative bright-field pictures of fish not injected (not inj.), zebrafish injected with snf8 MO with or without mRNA encoding wild-type SNF8, or the disease-associated SNF8Y167∗;G191D, SNF8P79L;V102I, SNF8V102I, or SNF8Y167∗ alleles at 48 hpf. The dashed vertical lines separate different images from different representative fields of view.
(B and C) Quantification of the percentage of fish showing normal (white) or aberrant (cyan) development upon injection of different snf8 MO doses (B) and of snf8 MO at 0.8 mM with or without the WT and mutant SNF8 mRNAs (C). In (B), n = 75 (not inj. on the left), 19 (snf8 MO 0.6 mM), 67 (snf8 MO 0.8 mM), and 104 (not inj. on the right). In (C) (left), n = 39 (not inj.), 8 (snf8 MO at 0.8 mM), 23 (snf8 MO + SNF8WT), 11 (snf8 MO + SNF8Y167∗;G191D), 18 (snf8 MO + SNF8P79L;V102I). In (C) (right), n = 165 (not inj.), 99 (snf8 MO at 0.8 mM), 98 (snf8 MO + SNF8WT), 55 (snf8 MO + SNF8V102I), 39 (snf8 MO + SNF8Y167∗).
(D–F) Representative confocal maximum intensity projections of fish fluorescently stained with antibodies against acetylated tubulin (cyan) and pH3 (magenta) from not injected (not inj.) (D), mild and severe cases of fish injected with snf8 MO (E), and fish co-injected with snf8 MO (at 0.8 mM) and SNF8WT (F). Fish injected with snf8 MO show a reduced anterior brain area and ectopic proliferative cells within the forebrain partially rescued by SNF8WT.
(G) Quantification of the overall brain area from the confocal z-projections.
(H) Number of embryos showing “normal” or “mildly or severely ectopic” proliferative cells (pH3+) within the anterior ventral forebrain. n = 15 (not inj. and snf8 MO) and 12 (snf8 MO + SNF8WT). Data are expressed as boxplot, for experiments including different batches mean ± SEM are shown.
Number of replicates in (B), one and two (left and right graphs, respectively). In (C) (left), number of replicates: three for not inj, snf8 MO, snf8 MO + SNF8WT, and snf8 MO + SNF8Y167∗;G191D; two for snf8 MO + SNF8P79L;V102I. In (C) (right), number of replicates: six (not inj, snf8 MO, and snf8 MO + SNF8WT); three (the other two mutants). Two-sided chi-square test is used to assess statistical significance in a 2×2 contingency table. In (B), not inj. vs. snf8 MO, ∗∗∗p = 0.0008 left and ∗∗∗∗p < 0.0001 right. In (C) (left), not inj. vs. snf8 MO, ∗∗∗p = 0.0005; snf8 MO vs. snf8 MO + SNF8WT, ∗p = 0.0312; snf8 MO + SNF8WT vs. + SNF8Y167∗;G191D, ∗∗p = 0.0086; snf8 MO + SNF8WT vs. snf8 MO + SNF8P79L;V102I, not significant (ns, p = 0.06); snf8 MO vs. mutants, ns. In (C) (right): not inj. vs. MO, ∗∗∗∗p < 0.0001; snf8 MO vs. snf8 MO + SNF8WT, ∗∗∗p = 0.0008; snf8 MO + SNF8WT vs. snf8 MO + SNF8V102I, ∗∗p = 0.0021; snf8 MO + SNF8WT vs. snf8 MO + SNF8Y167∗, ∗∗p = 0.0022 (ns: not significant). In (G) non-parametric Kruskal-Wallis test with Dunn’s post hoc test is used for statistical assessment (not inj. vs. snf8 MO, ∗p = 0.0141; snf8 MO vs. snf8 MO + SNF8WT, ∗∗p = 0.0032; ns = not significant). In (H), two-sided chi-square test is used: not inj. vs. snf8 MO, ∗p = 0.0309; not inj. vs. snf8 MO + SNF8WT, p = 0.8695 (ns, not significant); and snf8 MO vs. snf8 MO + SNF8WT, p = 0.0621 (ns). Fb, forebrain; oe, olfactory epithelium; OC, optic chiasm indicated by a white arrow. Asterisks indicate the eyes.
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Am. J. Hum. Genet.