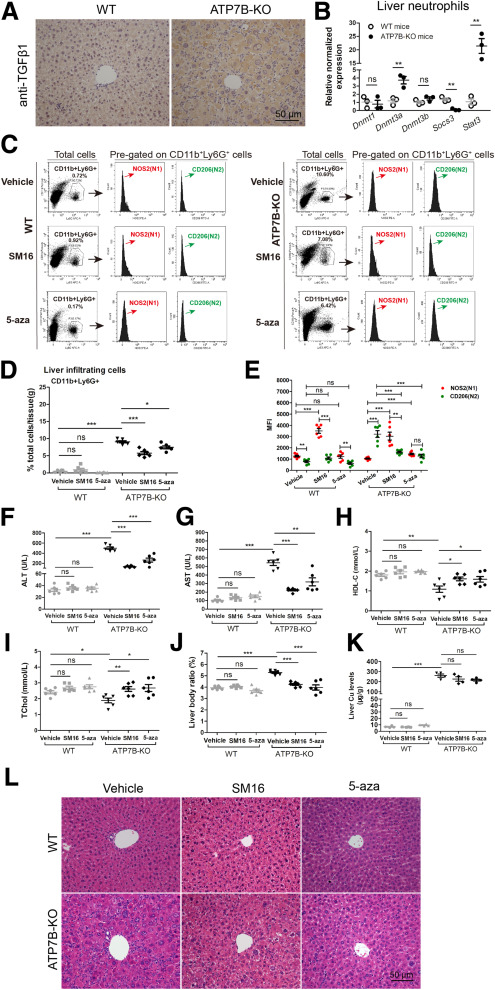
Fig. 13 Inhibition of TGFβ1 signaling or DNA methylation in ATP7B-KO mice decreases liver N2-neutrophil polarization and improves liver pathology. (A) Representative immunohistochemical staining images of anti-TGFβ1 in wild-type and ATP7B-KO mice livers. Scale bar: 50 μm. (B) qPCR of gene expression changes in Dnmt1, Dnmt3a, Dnmt3b, Scos3, and Stat3 in wild-type and ATP7B-KO mice liver neutrophils. n = 3 sets of neutrophils pooled from 2∼3 mice. ∗∗P < .01. (C) Flow cytometric analysis of N1 (CD11b+Ly6G+NOS2+) and N2 (CD11b+Ly6G+CD206+) neutrophil population in total CD11b+Ly6G+ cells derived from the livers of wild-type and ATP7B-KO mice treated with/without SM16 or 5-aza. (D) Flow cytometric quantification of CD11b+Ly6G+ neutrophils from wild-type and ATP7B-KO mice livers treated with/without SM16 or 5-aza. (E) Flow cytometric quantification of liver N1 and N2 neutrophils from wild-type and ATP7B-KO mice treated with/without SM16 or 5-aza. (D and E) n = 6 mice/group, unpaired 2-tailed t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. Serum activities of (F) ALT, (G) AST, (H) HDL-C, and (I) TChol in wild-type and ATP7B-KO mice treated with/without SM16 or 5-aza. n = 6 mice/group. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. (J) Liver body ratio in wild-type and ATP7B-KO mice treated with/without SM16 or 5-aza. n = 6 mice/group. ∗∗∗P < .001. (K) Liver Cu content in wild-type and ATP7B-KO mice treated with/without SM16 or 5-aza. n = 4 mice/group. ∗∗∗P < .001. (L) Representative H&E images in liver sections from wild-type and ATP7B-KO mice treated with/without SM16 or 5-aza. Scale bar: 50 μm. WT, wild-type.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Mol Gastroenterol Hepatol