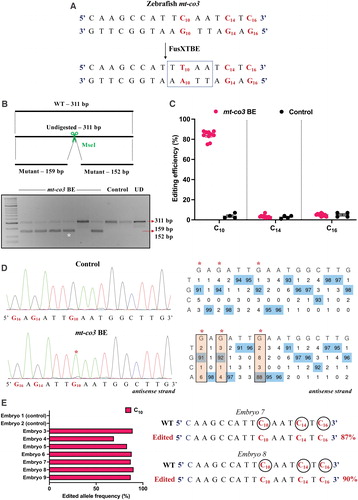
Fig. 7 Engineering precise base edits in vivo in the zebrafish mt-co3 gene. (A) Sequence of the potential editing sites in the mt-co3 protospacer region. The resulting gain of the restriction site for the enzyme MseI postediting event is marked by a dotted rectangle. (B) Screening of embryos harboring mutations by RFLP. C-to-T edits in the protospacer region lead to gain of recognition site for the MseI restriction enzyme. PCR amplicons from the control embryos show the presence of uncut product (311 bp). PCR amplicons from the injected embryos indicate the presence of expected 159 and 152 bp digested bands marked by a white asterisk (*). Each lane represents an individual embryo. (C) Editing efficiency for the target C10 present in the protospacer region in both groups of zebrafish embryos. Red and black color circles denote mutant and control embryos, respectively. Each data point represents an individual zebrafish embryo. Data are represented from independent experiments (Injected: N = 140 embryos and were screened and N = 10 embryos [RFLP positive] were genotyped by Sanger sequencing; Control = 60 embryos were screened and N = 4 embryos were genotyped by Sanger sequencing for which data have been included in the graph). (D) Representative chromatogram of the control and injected embryos. Asterisk (*) denotes the site of edit with corresponding editing percentage (C-to-T or G-to-A). Chromatograms and editing table plot were obtained using EditR. The combination of left arm- pT3-FusXTBE-N and right arm- pT3-FusXTBE-C was used to obtain the edits. (E) Percentage of reads with C-to-T transitions estimated by deep sequencing of target amplicon. Representative data from embryo 8 show the editing frequency of cytosine residues (C10, C14, and C16) in the protospacer region (highlighted in red and circled). The combination of left arm- pT3-FusXTBE-N and right arm- pT3-FusXTBE-C was used to obtain the edits. Color images are available online.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ CRISPR J