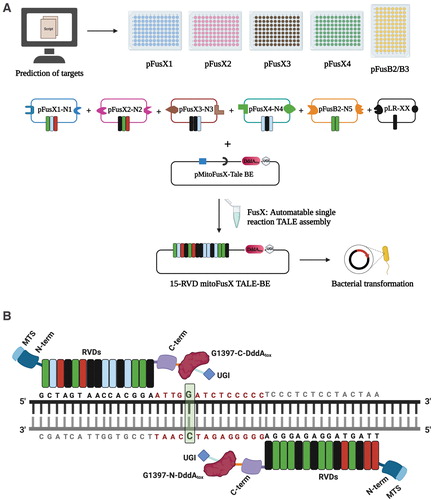
Fig. 1 Schematic for the assembly of mitochondrial TALE base editors by the FusX reaction with the FusXTBE architecture. (A) FusX library of all combinations of RVD dimers and trimers enables the mitochondrial TALE base editor to be built in a single Golden Gate cloning step. (B) Schematic of FusXTBE binding to the human mitochondrial DNA target, ND4. Repeat divariable residues are shown in different colors (green-guanine-NN; blue-cytosine-HD; red-thymine-NG; black-adenine-NI). DNA binding region for each of the TALE monomers is black in color, and the spacer region is depicted in red. Genomic target C is highlighted in green with bold font. Figure 1 was created using Biorender.com. FusXTBE, FusX TALE Base Editor; MTS, mitochondrial targeting sequence; RVD, repeat variable di-residues; UGI, uracil glycosylase inhibitor. Color images are available online.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ CRISPR J