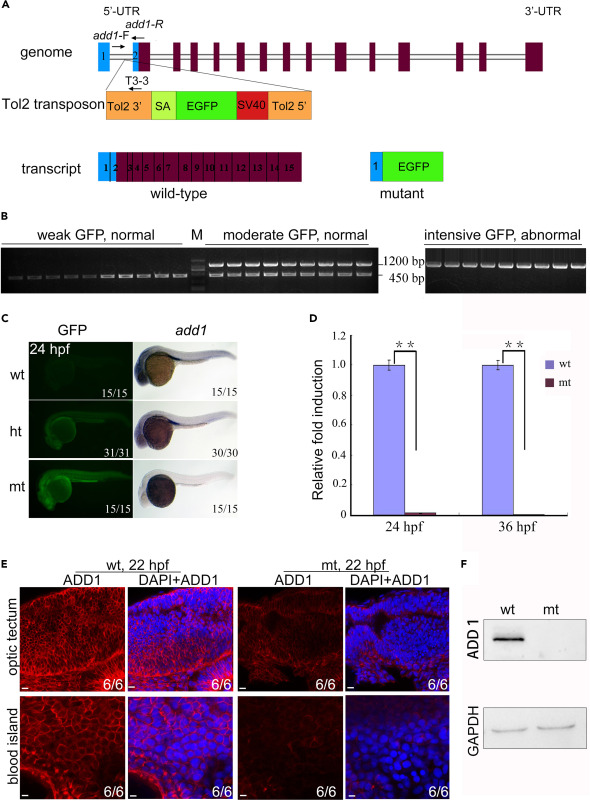
Fig. 2 Genomic schematic and reduced expression level of add1 in add1−/− mutant embryos (A) Genomic structure of the add1 locus and putative transcripts. The Tol2 transposon was inserted into the first intron of the add1 gene. The binding positions and directions of primers add1-F, add1-R, and T3-3 used in (B) were indicated. UTR, untranslated region. (B) PCR-based genotyping of individual embryos. At 24 hpf, embryos from add1 heterozygote intercrosses were separated based on GFP intensity and phenotypes. Three primers, add1-F, add1-R, and T3-3, the Tol2-3′ element located downstream of the insertion site, were used in tandem for PCR with genomic DNA from a single embryo as the template. The mutant and WT add1 alleles were represented by 1200 and 450 bp bands, respectively. M stands for molecular marker lane. (C) Expression levels of add1 transcript in mutant embryos detected with WISH. In the left panel, wt, ht, and mt represented WT with weak GFP, heterozygote with moderate GFP, and homozygote with intensive GFP, respectively. It is worth noting that the expression level of Add1 nearly vanished. (D) qRT-PCR analysis of the relative expression level of add1 in WT and add1−/− embryos at indicated stages. β-actin was used as an internal control. Embryos were pre-sorted as mentioned above. The data were presented as mean ± SD from three independent biological repeats. Total RNA of each group was extracted from a pool of 30 embryos. ∗∗, p < 0.01 (by Student’s t test). (E and F) Expression of Add1 was dramatically reduced in add1−/− mutants. Embryos of add1−/− and their siblings were harvested at 22 hpf, and then stained with anti-Add1 antibodies. Confocal photography was used to capture the optic tectum and blood island. (E). Scale bars, 10 μm. The levels of Add1 in cell lysis of the embryos was further analyzed by western blots (F).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ iScience