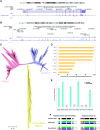
Fig. 3 Functional diversification of PKNOX genes in vertebrates. (A) PKNOX1 locus in chromosome 21 and (B) PKNOX2 locus in chromosome 11 of the human genome (GRCh37/hg19). (C) Phylogenetic gene tree reconstruction of PKNOX proteins across vertebrate evolution, using hierarchical orthologous groups (HOGs) from the OMA Browser (https://omabrowser.org/). The evolutionary history was inferred using the neighbor-joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix–based method and are in the units of the number of amino acid substitutions per site. The analysis involved 218 amino acid sequences. (D) Proportion and (E) total number of coding and noncoding conserved elements (PhastCons) in TALE proteins. (F) Functional diversification hypothesis that could explain the regulatory domain gain and loss that resulted into the vertebrate PKNOX gene expression pattern.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Mol Bio Evol