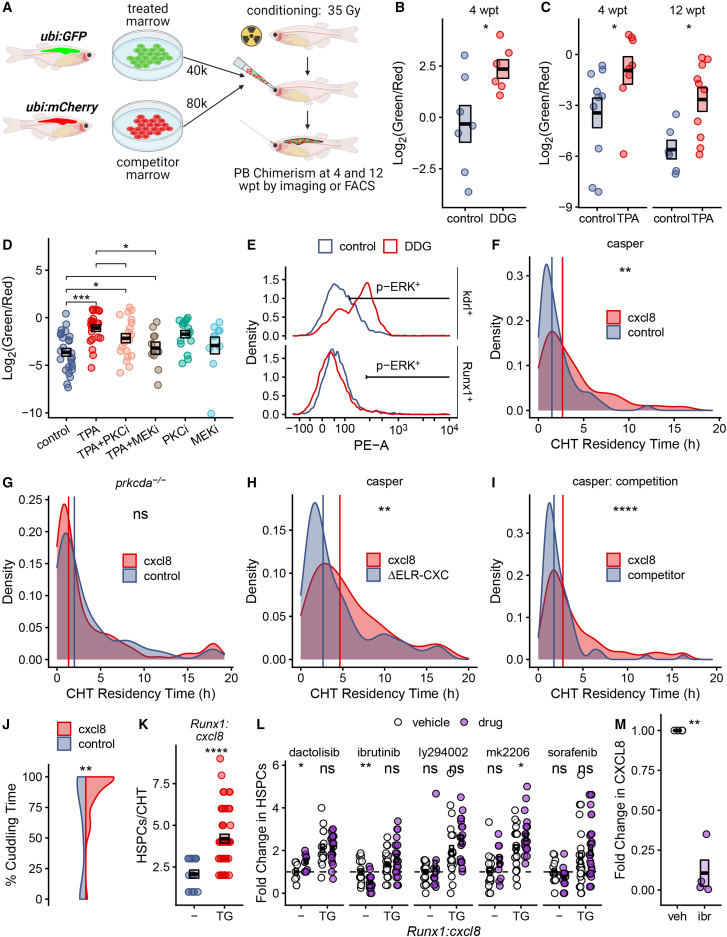
Fig. 5 PKC agonists augment HSPC engraftment and niche colonization in competition assays (A) Schematic illustration of competitive marrow transplant experiments. (B–D) Competitive transplant data are presented as Log2(green/red) where green/red is the calculated ratio of drug- or control-treated marrow to untreated competitor marrow. Boxes represent mean ± SEM; each point is a biological replicate. (B) DDG vs. vehicle control at 4 wpt: DDG:competitor ratio 6.58 ± 2.18 vs. control:competitor ratio 2.11 ± 1.12, p = 0.026, Welch’s t test. (C) TPA vs. vehicle control at 4 wpt: TPA:competitor ratio 0.99 ± 0.31 vs. control:competitor ratio 0.24 ± 0.07, p = 0.027, Wilcoxon rank-sum test. 12 wpt: TPA:competitor ratio 0.34 ± 0.11 vs. control:competitor ratio 0.03 ± 0.01, p = 0.016, Wilcoxon rank-sum test. (D) TPA treatment in the presence or absence of PKC and MEK inhibitors, vs. control. TPA vs. TPA+MEKi: TPA:competitor ratio 0.71 ± 0.13 vs. TPA+MEKi:competitor ratio 0.22 ± 0.08, p = 0.018, Wilcoxon rank-sum test. (E) ERK phosphorylation was assessed by intracellular flow cytometry in sorted kdrl+ endothelial cells and Runx1+ HSPCs. (F) Distribution of CHT residency times for n = 46 HSPCs from Runx1:cxcl8 transgenics and n = 62 HSPCs from control clutchmates. Vertical lines indicate the median for each group. Representative data from one of four similar experiments is shown. For cxcl8 vs. control: median of 2.67 vs. 1.5 h, p = 0.003, Wilcoxon rank-sum test. (G) Distribution of CHT residency times for Runx1:cxcl8 transgenics and control clutchmates in the prkcda(−/−) mutant background. n = 108 HSPCs for Runx1:cxcl8 and n = 111 HSPCs for control in 10 fish per group. (H) Distribution of CHT residency times for n = 110 HSPCs from Runx1:cxcl8 transgenics and n = 223 HSPCs from ΔELR-CXC clutchmates. Vertical lines indicate the median for each group. For cxcl8 vs. ΔELR-CXC; median of 4.66 vs. 2.66 h, p = 0.002, Wilcoxon rank-sum test. (I) Distribution of CHT residency times for n = 404 HSPCs for Runx1:cxcl8 and n = 174 HSPCs for competitor. Vertical lines indicated the median for each group. For cxcl8 vs. competitor: median of 2.75 vs. 1.75 h, p = 1 × 10−9, Wilcoxon rank-sum test. (J) Violin plots represent the percentage of time individual HSPCs were cuddled by endothelial cells within the CHT. n = 32 HSPCs for Runx1:cxcl8 and n = 108 HSPCs for control. For cxcl8 vs. control: median of 97.7% vs. 79.3% cuddling time, p = 0.0013, Wilcoxon rank-sum test. (K) Quantification of Runx1:EGFP+ HSPCs in Runx1:EGFP;Runx1:cxcl8-2A-mCherry transgenics (TG) and Runx1:EGFP clutchmates (−). Data are from one of two representative zebrafish clutches. Boxes represent mean ± SEM; each point is a biological replicate. For TG vs. −: 4.2 ± 0.27 vs. 2.1 ± 0.25 HSPCs per CHT, p = 5.5 × 10−7, Welch’s t test. (L) Quantification of Runx1:EGFP(+) HSPCs in Runx1:cxcl8-2A-mCherrytransgenics (TG) and cxcl8 transgene-negative control clutchmates (−) treated with the indicated drugs or vehicle control. Data are presented as fold change relative to control-treated, transgene-negative clutchmates and are pooled from eight independent experiments. Boxes represent mean ± SEM; each point is a biological replicate. For ibrutinib-treated cxcl8 transgene-negative zebrafish: 0.51 ± 0.08-fold relative to vehicle-treated clutchmates, p = 0.0067, Welch’s t test. (M) Quantitative RT-PCR for CXCL8 mRNA expression by THP-1 cells treated with ibrutinib or vehicle control. Four biological replicates were performed per condition; each point is the mean of three technical replicates. For ibrutinib-treated cells: 0.11 ± 0.08-fold decrease relative to vehicle control, p = 0.002, Welch’s t test. See also Figure S4.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.