Fig. 7
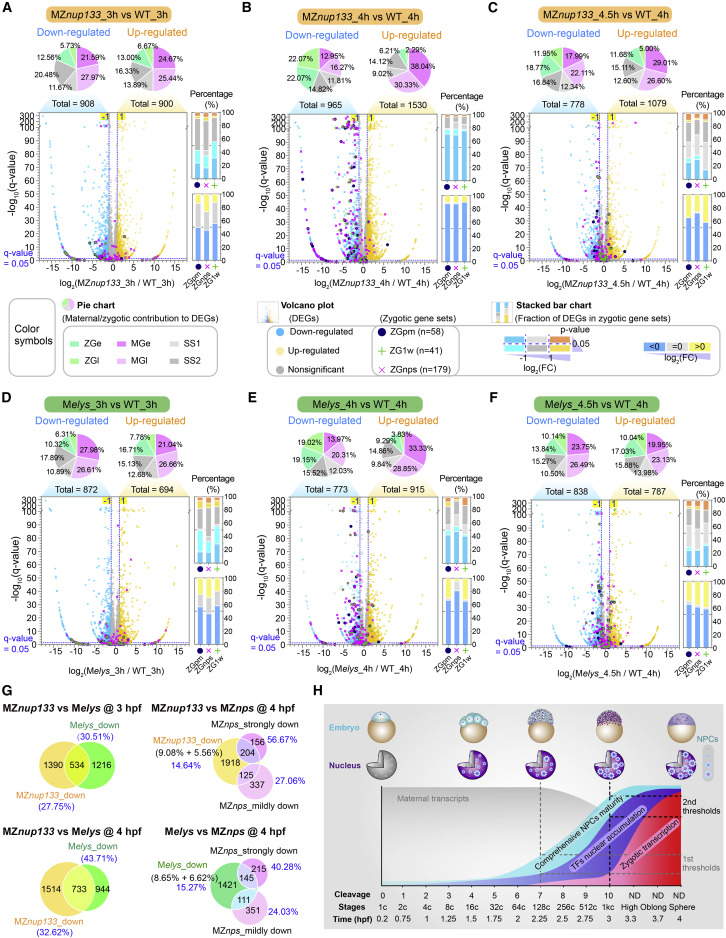
Figure 7. Impact of nup133 or elys mutation and nup133 overexpression on ZGA (A–F) Transcriptome comparison between MZnup133 (A–C) or Melys (D–F) mutant and WT embryo at different stages. Top panel, composition of differentially expressed genes (DEGs) with fold change >2 or <0.5 and q value < 0.05. Gene classes60: ZGe, early zygotic genes; ZGl, late zygotic genes; MGe, early degraded maternal genes; MGl, late degraded maternal genes; SS1, semi-stable I genes; SS2, semi-stable II genes. Bottom panel, zygotic gene classes: ZGpm, pre-MBT zygotic-only genes18,23; ZG1w, first-wave zygotic-only genes18; ZGnps, zygotic genes strongly downregulated in MZnanog;MZpou5f3;MZsox19b triple mutants (MZnps) at 4 hpf.26 The number of zygotic genes within a class was listed in parenthesis. (G) Venn diagram showing proportions of downregulated genes shared between different types of mutants. (H) The model of clock-like comprehensive NPCs maturity for ZGA timing control. The time point for minor or major ZGA was indicated by a vertical gray or black dotted line, respectively. NPCs in 1c-stage embryo were colored in gray without experimental examination. ND, not determinable due to asynchronous cell divisions. See also Figures S5 and S6 and Table S2.
Reprinted from Cell, 185(26), Shen, W., Gong, B., Xing, C., Zhang, L., Sun, J., Chen, Y., Yang, C., Yan, L., Chen, L., Yao, L., Li, G., Deng, H., Wu, X., Meng, A., Comprehensive maturity of nuclear pore complexes regulates zygotic genome activation, 4954-4970.e20, Copyright (2022) with permission from Elsevier. Full text @ Cell