Figure 2
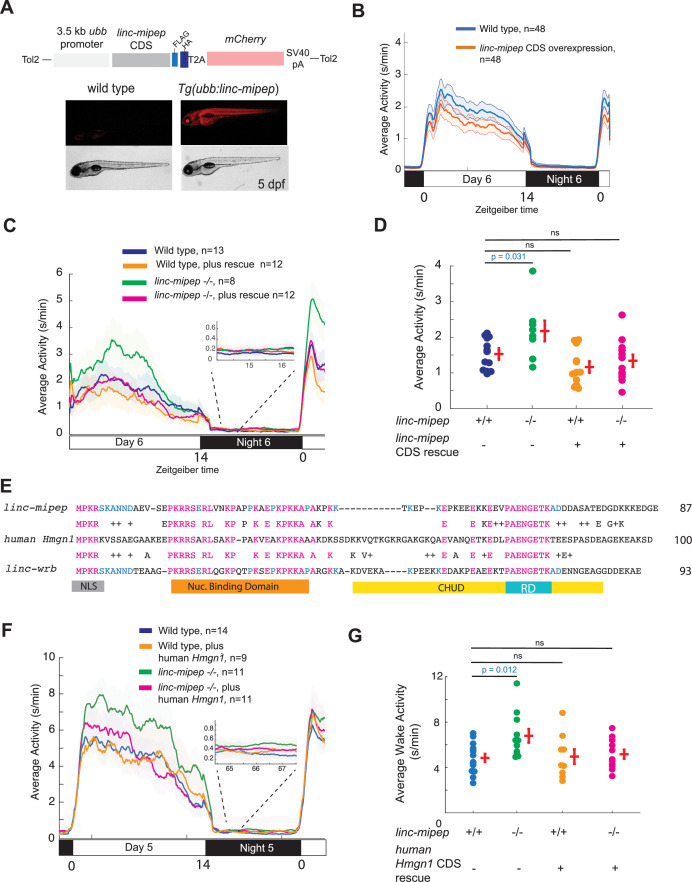
(A) Top, diagram of transgenic linc-mipep overexpression construct. Transgenic lines were established via Tol2-mediated integration of 3.5kb ubiquitin B (ubb) promotor driving the linc-mipep coding sequence with a FLAG and HA tag at the C-terminus, followed by a T2A self-cleaving peptide, mCherry reporter, and SV40 polyA tail. Bottom, fluorescent and brightfield images of 5 dpf zebrafish siblings either without overexpression (wild type, mCherry-negative, left) or with linc-mipep overexpression (mCherry-positive, right). (B) Activity plot of wild type (mCherry-negative, blue) or linc-mipep overexpression (Tg(ubb:linc-mipep) mCherry-positive, orange) siblings at 6 dpf. n=48 per genotype. Average day activity p=0.028, one-way ANOVA. (C) Locomotor activity of linc-mipep mutants, with or without transgenic linc-mipep overexpression (Tg(ubb:linc-mipep CDS-T2A-mCherry), ‘rescue’), sibling-matched larvae over 24hr. Inset, no effect on nighttime activity. (D) Average waking activity of 6 dpf linc-mipep mutant, heterozygous, or wild type larvae, with (denoted by +) or without (denoted by -) linc-mipep transgenic rescue. Each dot represents a single fish, and crossbars plot the mean ± SEM. p Values from a Dunnett’s test, using wild type (linc-mipep +/+) as the baseline condition. (E) Amino acid sequences of linc-mipep (top), linc-wrb (bottom), and human Hmgn1 (middle). Conserved amino acids are denoted in blue (if conserved between two sequences) or magenta (if conserved across the three sequences). Conserved functional domains for Hmgn1 are denoted (NLS, nuclear localization signal; Nuclear Binding Domain; RD, Regulatory Domain; and CHUD, Chromatin Unwinding Domain). (F) Locomotor activity of linc-mipep mutants, with or without transgenic human Hmgn1 overexpression (Tg(ubb:hHmgn1CDS-T2A-mCherry), ‘rescue’), sibling-matched larvae over 24hr. Inset, no effect on nighttime activity. (G) Average waking activity of 6 dpf linc-mipep mutant (-/-) or wild type (+/+) larvae, with (denoted by +) or without (denoted by -) human Hmgn1 transgenic rescue. Each dot represents a single larva, and crossbars plot the mean ± SEM. p Values from a Dunnett’s test, using wild type (linc-mipep +/+) as the baseline condition.
linc-mipep and linc-wrb encode proteins with homology to human HMGN1.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife