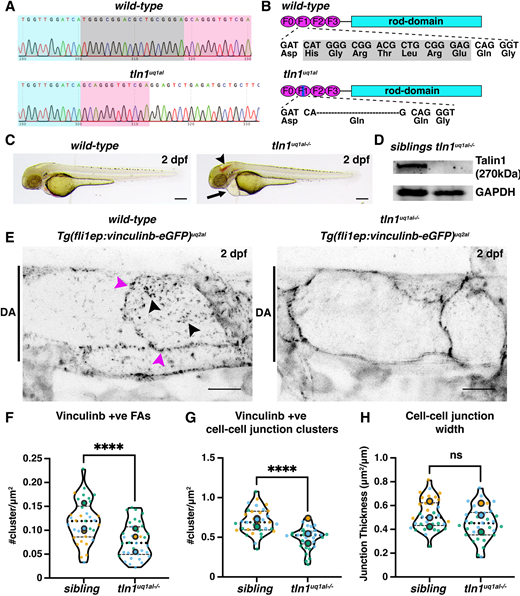
Fig. 1
Novel zebrafish models for studying focal adhesion function and dynamics in live vasculature. (A) Chromatograms of genomic sequencing of a wild-type sibling embryo (top) and a homozygous tln1uq1al−/− mutant (bottom). Bases highlighted in grey in the wild type are deleted in tln1uq1al−/− mutants. (B) Schematics of Talin 1 protein showing the 18 bp deletion in tln1uq1al−/− resulting in an in-frame deletion of six amino acids (His-Gly-Arg-Thr-Leu-Arg) and a Glu-to-Gln substitution in the F1 FERM domain. (C) Bright-field images showing body morphology of a wild-type sibling versus a tln1uq1al−/− mutant at 2 dpf. Arrowhead indicates cranial haemorrhaging in the mutant. Arrow indicates cardiac oedema in the mutant. Scale bars: 100 µm. (D) Western blot analysis at 3 dpf showing loss of Talin 1 in tln1uq1al−/− mutants. (E) Expression of Vinculinb-eGFP at FAs (black arrowheads) and at cell-cell junctions (magenta arrowheads) in the dorsal aorta (DA) of a wild-type sibling at 2 dpf (left). In tln1uq1al−/− mutants (right), expression at FAs in greatly reduced, while a diffuse presence of Vinculinb-eGFP is observed at cell-cell junctions. Scale bars: 10 µm. (F) Number of Vinculinb-eGFP-positive FA clusters, n=3 replicates: n=23 ECs siblings and n=18 ECs tln1uq1al−/− mutants (****P<0.0001, unpaired t-test ). (G) Number of Vinculinb-eGFP-positive clusters at cell-cell junctions, n=3 replicates: n=23 ECs siblings and n=18 ECs tln1uq1al−/− mutants (****P<0.0001, unpaired t-test ). (H) Quantification of cell-cell junction width based on Vinculin-eGFP expression, n=3 replicates: n=23 ECs siblings and n=18 ECs tln1uq1al−/− mutants, no significant difference (ns) (unpaired t-test). Replicate averages are depicted by large circles; smaller circles indicate individual data points of each replicate (colour matched).