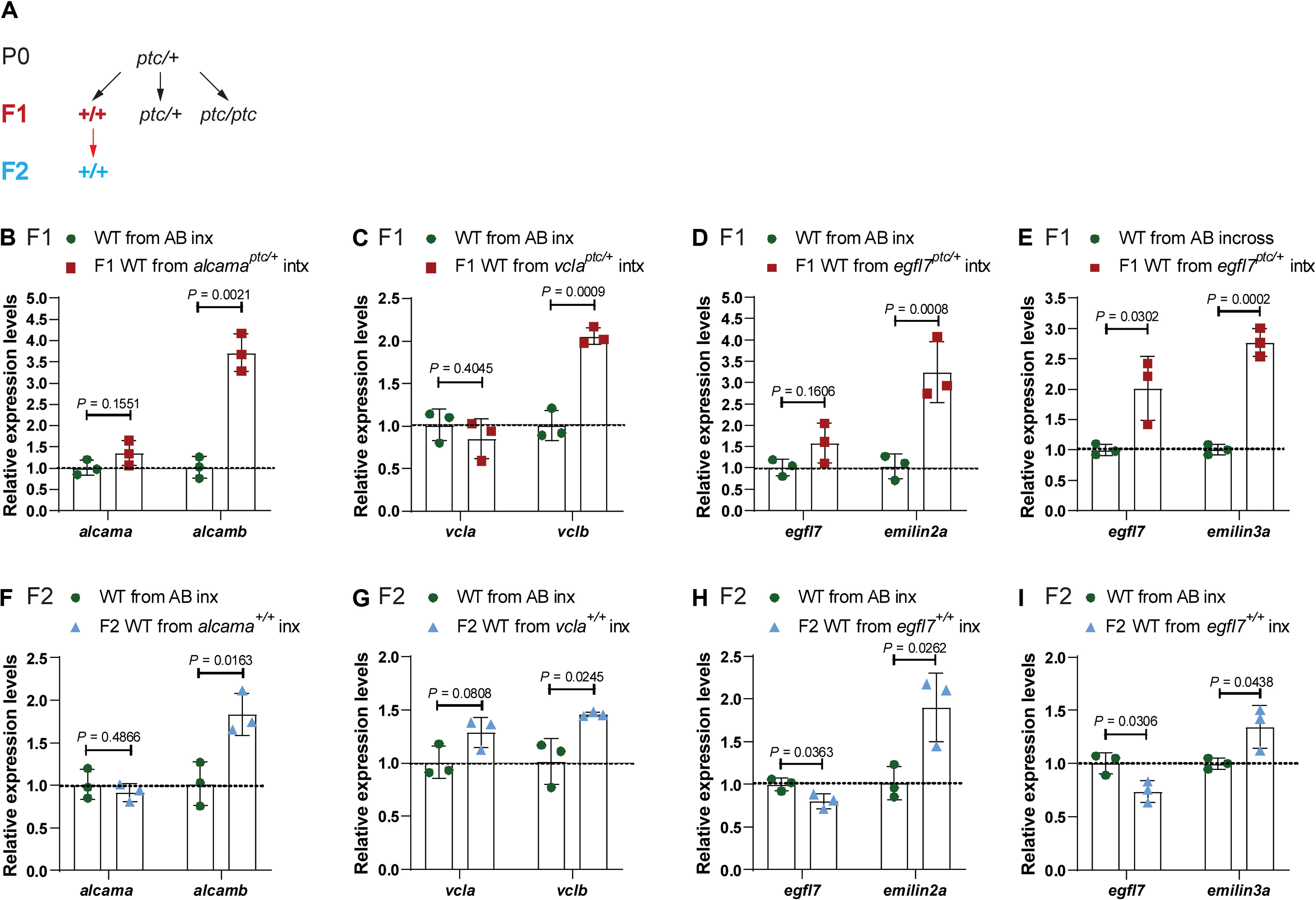
Fig. 2 (A) Genetic crosses used to obtain wild-type (WT) embryos from TA-displaying heterozygous zebrafish and from subsequent incrosses. Black arrows indicate heterozygous intercrosses (intx); red arrow indicates subsequent wild-type incrosses (inx). (B) Relative mRNA levels of alcama and alcamb in 28 hpf wild-type embryos from AB incrosses and from alcamaptc/+ intercrosses. (C) Relative mRNA levels of vcla and vclb in 24 hpf wild-type embryos from AB incrosses and vclaptc/+ intercrosses. (D) Relative mRNA levels of egfl7 and emilin2a in 24 hpf wild-type embryos from AB incrosses and egfl7ptc/+ intercrosses. (E) Relative mRNA levels of egfl7 and emilin3a in 24 hpf wild-type embryos from AB incrosses and egfl7ptc/+ intercrosses. (F) Relative mRNA levels of alcama and alcamb in 28 hpf wild-type embryos obtained from AB incrosses and F1 alcama+/+ (i.e., wild types from alcamaptc/+ intercrosses) incrosses. (G) Relative mRNA levels of vcla and vclb in 24 hpf wild-type embryos obtained from AB incrosses and F1 vcla+/+ (i.e., wild types from vclaptc/+ intercrosses) incrosses. (H) Relative mRNA levels of egfl7 and emilin2a in 24 hpf wild-type embryos obtained from AB incrosses and F1 egfl7+/+ (i.e., wild types from egfl7ptc/+ intercrosses) incrosses. (I) Relative mRNA levels of egfl7 and emilin3a in 24 hpf wild-type embryos obtained from AB incrosses and F1 egfl7+/+ (i.e., wild types from egfl7ptc/+ intercrosses) incrosses. n = 3 biologically independent samples. Control expression levels were set at 1. Data are means ± SD, and a two-tailed Student’s t test was used to calculate P values. Ct values are listed in table S1.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Sci Adv