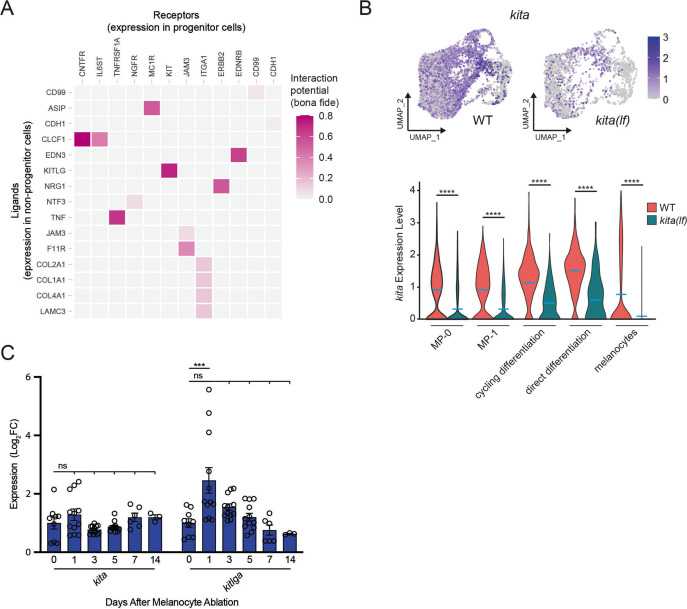
Figure 3
(A) Heatmap of NicheNetR-identified ligand/receptor pairs indicating interaction potential between receptors on progenitor receiver cells and ligands on non-progenitor sender cells sampled by scRNAseq. Ligand/receptor pairs were restricted to literature-supported pairs (Browaeys et al., 2020). (B) Top, feature and, bottom, violin plots of kita expression in mitfa+aox5lo cells from wild-type (MP-0 = 1840, MP-1 = 1408, cycling differentiation = 777, direct differentiation = 535, and melanocytes = 1060 cells) and kita(lf) strains (MP-0 = 617, MP-1 = 612, cycling differentiation = 280, direct differentiation = 57, and melanocytes = 563 cells). Mean gene expression represented by cyan bars. p values calculated by Wilcoxon rank-sum test, ****p < 0.0001. (C) Quantitative real-time polymerase chain reaction (qRT-PCR) of kita and kitlga expression in zebrafish skin following melanocyte destruction. Three biological replicates were performed for each time point. Data are shown as mean ± standard error of the mean (SEM). p values calculated by one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test, ***p < 0.001; ns, not significant.
NicheNet and transcriptional analyses implicate the KIT signaling axis as a dynamic regulator of melanocyte regeneration.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife