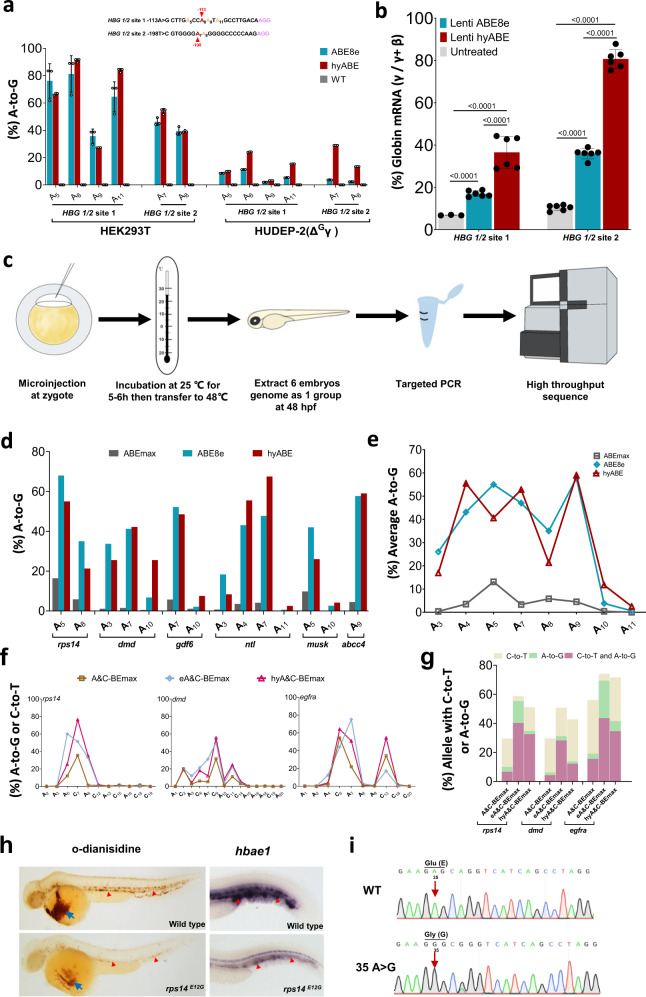
Fig. 4
a The comparison of base editing efficiency by ABE8e and hyABE at HBG 1/2 −113 A-to-G or −198 T-to-C in HEK293T cells and HUDEP-2 (ΔGγ) cells. Data are means ± SD (n = 3 independent experiments). b Comparison of γ-globin mRNA expression relative to β-like globin mRNA via hyABE treatment in HUDEP-2 (ΔGγ) cells after differentiation. Data are means ± SD (n = 3 for one untreated group and n = 6 for the other untreated or treated group). P value was determined by two-tailed Student’s t-test. c The flow chart for generating zebrafish mutants with hyABE and hyA&C-BEmax. d The A-to-G base editing efficiency of hyABE, ABE8e or ABEmax at endogenous target sites for ntl, dmd, musk, rps14, gdf6 and abcc4 in zebrafish embryos. e The average A-to-G editing efficiency of hyABE, ABE8e, or ABEmax across protospacers at the endogenous targets shown in b. f The A-to-G or C-to-T editing efficiency of A&C-BEmax, eA&C-BEmax or hyA&C-BEmax was examined at 3 endogenous target sites in zebrafish embryo. g The composition of A&C-BEmax, eA&C-BEmax and hyA&C-BEmax base editing products at 3 endogenous zebrafish genomic loci. The individual data are shown as yellow (only C-to-T), green (only A-to-G) and plum purple (simultaneous C-to-T and A-to-G) columns. h The hematopoietic phenotypes of rps14E12G embryos. O-dianisidine staining exhibits a reduction of erythrocytes at the yolk sac (blue arrow) and the trunk region (red triangle) in rps14E12G embryos compared to wild-type (WT) control embryos at 48 hpf (left). WISH analysis for hbae1 expression (red triangle) in rps14E12G mutant embryos and WT control embryos at 72 hpf (right). i Sanger sequencing chromatograms of DNA from a WT embryo and a single F0 rps14E12G embryo carrying the A to G conversion at the nucleotide 35 (arrow) that causes Glu (E) to Gly (G) substitution (Red arrow). Source data are provided as a Source Data file.