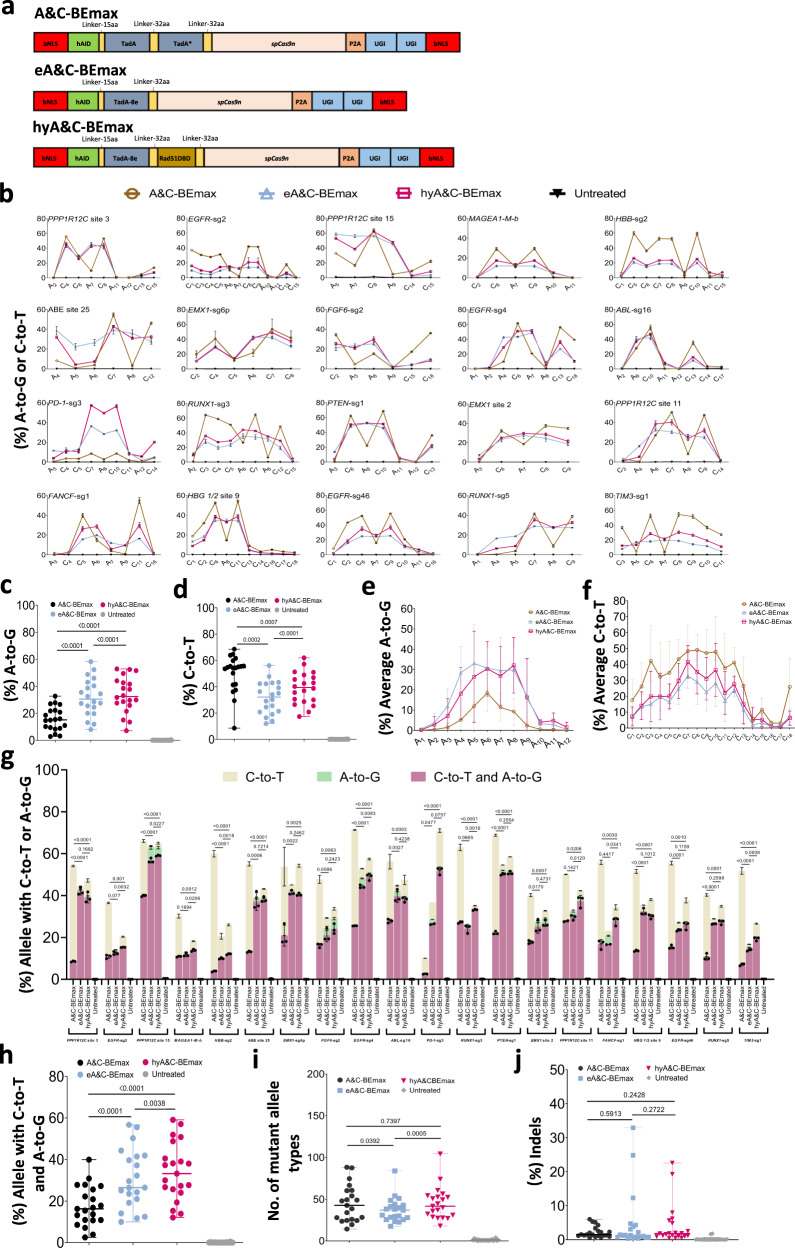
Fig. 2
a Schematics of the constructs with Rad51DBD fused to A&C-BEs. b The A-to-G or C-to-T editing efficiency of A&C-BEmax, eA&C-BEmax or hyA&C-BEmax was examined at 20 endogenous target sites in HEK293T cells. Data are means ± SD (n = 3 independent experiments). c Summary of the A-to-G editing efficiencies for only the most highly edited adenine induced by dual base editor at the 21 target sites in b and Supplementary Fig. 3b. d Summary of the C-to-T editing efficiencies for only the most highly edited cytosine induced by dual base editors at the 21 target sites in b and Supplementary Fig. 3b. e Average A-to-G editing efficiency of A&C-BEmax, eA&C-BEmax or hyA&C-BEmax at the same 21 target sites in b and Supplementary Fig. 3b. Data are means ± SD (n = 3 independent experiments). f Average C-to-T editing efficiency of A&C-BEmax, eA&C-BEmax, or hyA&C-BEmax at the same 21 target sites in b and Supplementary Fig. 3b. Data are means ± SD (n = 3 independent experiments). g The allele with A-to-G or C-to-T efficiency of A&C-BEs at 21 endogenous target sites in HEK293T cells. Data are means ± SD (n = 3 independent experiments). h Frequency of A/C simultaneous conversion induced by A&C-BEmax, eA&C-BEmax, and hyA&C-BEmax at 21 endogenous genomic loci in b and Supplementary Fig. 3b. i Number of mutant allele types formation by A&C-BEmax, eA&C-BEmax or hyA&C-BEmax at 21 endogenous genomic loci in b and Supplementary Fig. 3b. j Frequency of indels formation by A&C-BEmax, eA&C-BEmax or hyA&C-BEmax at 21 endogenous genomic loci in b and Supplementary Fig. 3b. For c, d, Each data point represents means at indicated target sites from 3 independent experiments. For h–j, Each data point represents means at indicated target sites calculated from 3 independent experiments. Significance was tested with two-tailed Student’s t test (e–g) or two-sided paired Wilcoxon rank-sim test (c, d and h–j). Source data are provided as a Source Data file.