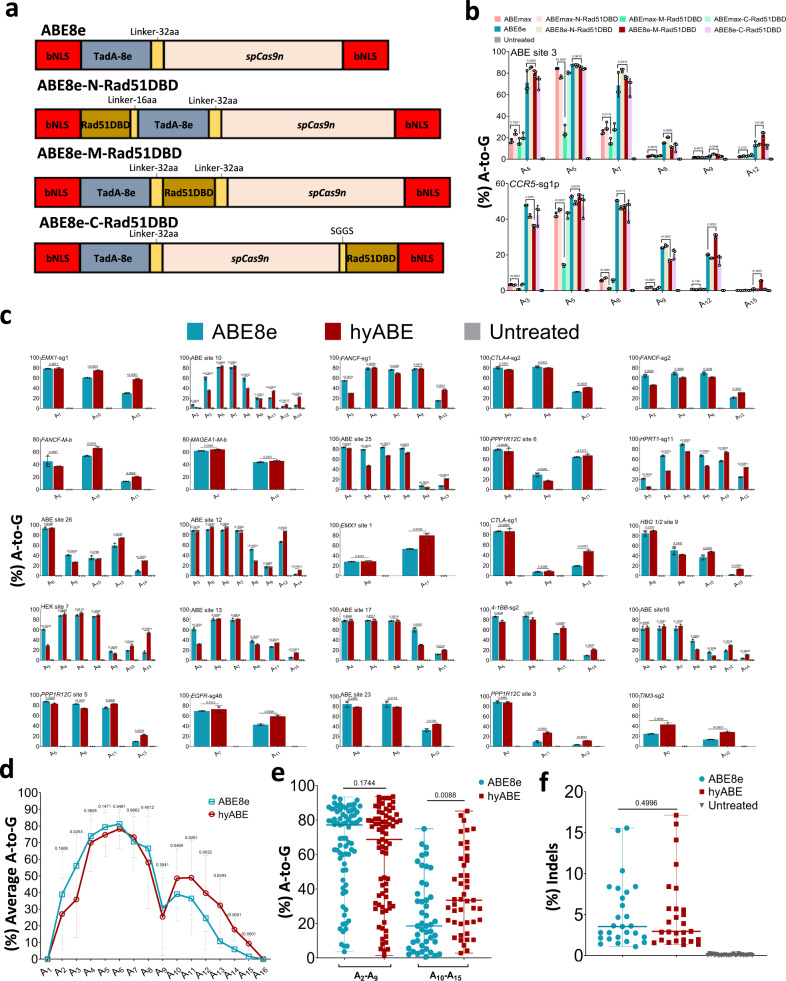
Fig. 1
a Schematics of the constructs with Rad51DBD fused to ABE8e. bNLS, bipartite nuclear localization signals; TadA-8e, derived from evolved E.coil adenosine deaminase; spCas9n, Cas9 D10A; Rad51DBD, single-strand DNA binding domain; Linkers are also shown. b The A-to-G base editing efficiency of ABE8e or its fusion constructs at 2 endogenous genomic loci containing multiple As (ABE site 3 and CCR5-sg1p) in HEK293T cells. Data are means ± SD (n = 3 independent experiments). c The A-to-G editing efficiency of ABE8e or hyABE was examined at 25 endogenous genomic loci containing multiple As in HEK293T cells. Data are means ± SD (n = 3 independent experiments). d Average A-to-G editing efficiency of ABE8e or hyABE at each position within the protospacer across the 27 target sites in b and c. e Summary of the A-to-G editing efficiency for A2-A9 or A10-A15 induced by ABE8e or hyABE at the 27 target sites in b and c. Each data point represents average A-to-G editing efficiency at all As within the activity window of each target site, calculated from 3 independent experiments. f Frequency of indels formation by ABE8e and hyABE at 27 endogenous genomic loci in b, c. Each data point represents average indels frequency at each target site calculated from 3 independent experiments. Significance was tested with two-tailed Student’s t test (b–d) or two-sided paired Wilcoxon rank-sum test (e, f). Source data are provided as a Source Data file.