Fig. 5
PHLPP1-dependent neutral lipid accumulation involves ChREBP and AMPK
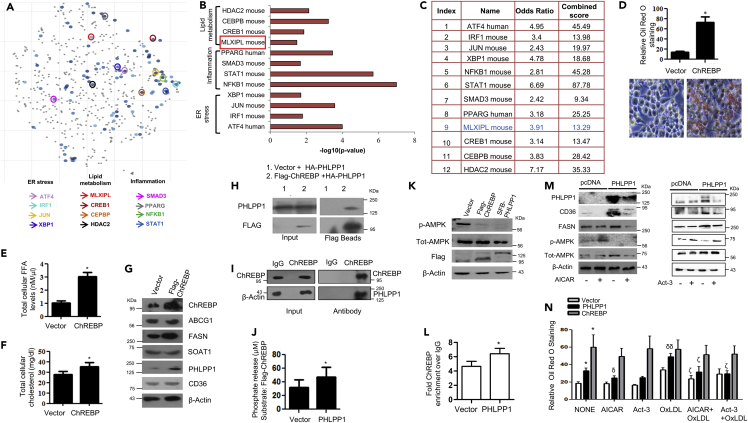
(A) Scatterplot of enriched transcription factors in set 3: Differentially altered gene-set from set 3 (cut-off: FC > 1.5, FDR 0.05) was analyzed by “Enrichr” tool to analyze the possible transcription factors involved in the gene alterations using ChIP-X enrichment analysis (ChEA) gene set library (The brighter the color, the more significant the transcription factor).
(B and C) Plot and table indicating the –log10 p value (B) and terms odds ratio, and combined score (C) used by the Enrichr software to represent the output (scatterplot). p values are computed based on alterations in the number of genes that are known primary or secondary targets of transcription factors; Odds ratio indicates a measure of association between input and outcome; combined score: log(p) ∗z, p is p value, and z = deviation from the expected rank.
(D) Neutral lipid staining by Oil Red O in ChREBP overexpressing cells along with quantification (top); Images are taken at |×20×| magnification. Each image is a representative image of four different experiments with a minimum of eight different fields in each experiment. Statistical analysis was performed using two-tailed t-test (∗p < 0.05, vs Vector).
(E and F) Total cellular FFA (E) and cholesterol levels (F) upon ChREBP overexpression. Statistical analysis was performed using two-tailed t-test (∗p < 0.05, vs Vector).
(G) Protein levels of foam cell markers in ChREBP overexpressing cells. Densitometric analysis is shown in Figure S5A.
(H and I) Pull-down assay: HA-PHLPP1 and Flag-ChREBP interaction in HEK293T cells (H) and endogenous interaction (I) between PHLPP1 and ChREBP in RAW 264.7 cells;
(J) Malachite green assay: phosphate release upon the incubation of Flag-ChREBP with SFB-PHLPP1 in vitro. Statistical analysis was performed using two-tailed t-test (∗p < 0.05, vs Vector).
(K) AMPK Thr172 phosphorylation in ChREBP or PHLPP1 overexpressing RAW 264.7 cells. Densitometric analysis is shown in Figure S5B.
(L) Graph showing enriched ChREBP binding to FASN promoter upon PHLPP1 overexpression in ChIP-qPCR. Statistical analysis was performed using two-tailed t-test (∗p < 0.05 vs vector).
(M) Effect of AICAR and Act-3 on CD36 and FASN protein levels in PHLPP1 overexpressing cells. Densitometric analysis is shown in Figures S5C and S5D.
(N) Oil Red O quantification of neutral lipids in PHLPP1 and ChREBP over-expressed RAW 264.7 cells treated with AICAR and Act3. Images from three different experiments with a minimum of eight different fields in each experiment were used to quantify the data. Statistical analysis was performed using one-way ANOVA followed by Bonferroni′s post hoc analysis (∗p < 0.05 versus vector-untreated; δp < 0.05, δδp < 0.01 versus corresponding control-untreated; ζp < 0.05 versus corresponding sample OxLDL-treated). Data are a representative of three independent experiments. Numerical data are expressed as mean ± SEM. See also Figures S4 and S5.