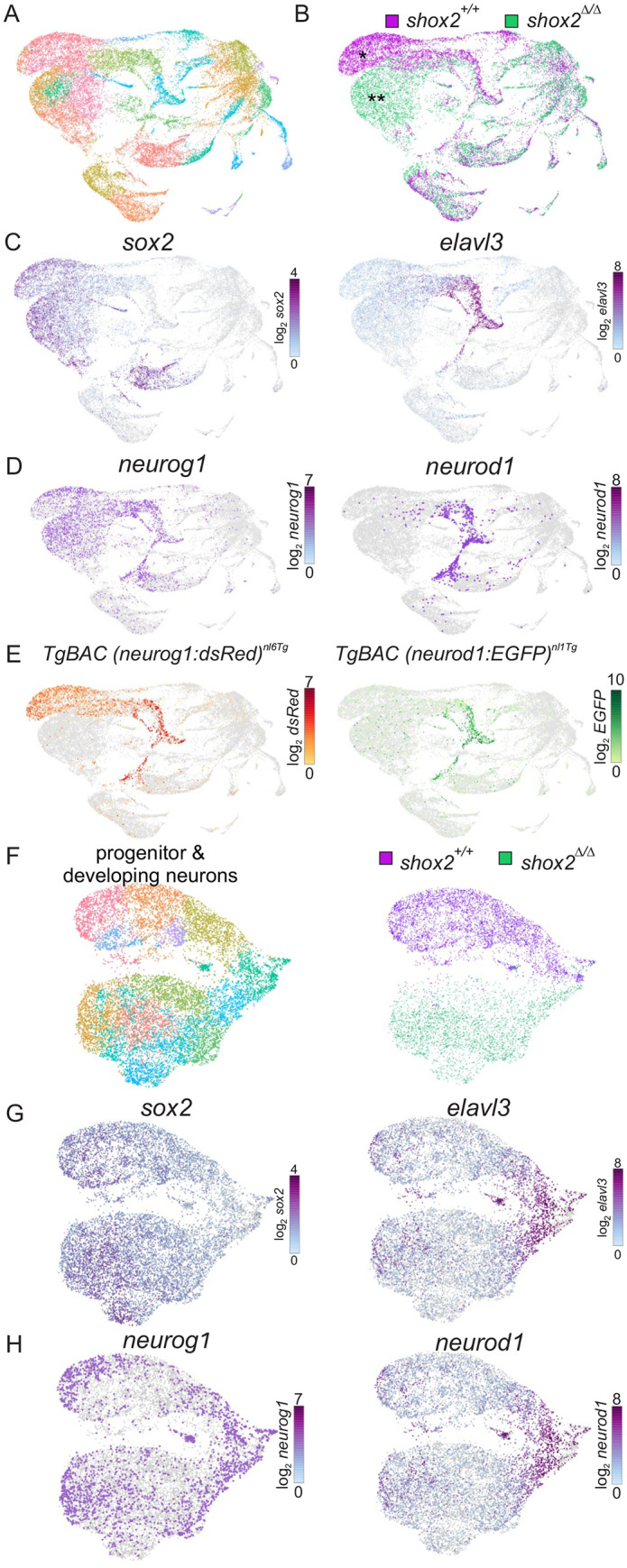
Fig. 6. Identifying cell populations in shox2+/+ and shox2Δ/Δ embryos by single cell RNA-seq. (A) shox2+/+ (n=59 embryos) and shox2Δ/Δ embryos (n=43 embryos) at 16-22 hpf were harvested and tissue around the otic vesicle microdissected. Samples of the same genotype were pooled and subjected to scRNA-seq. Transcriptome of individual cells were aggregated, dimensionality reduction performed followed by unsupervised clustering before visualizing as a UMAP projection. Course-grain visualization of 29,152 cells identified 28 distinct cell types after clustering. (B) Cells from shox2+/+ (magenta) and shox2Δ/Δ (green) were identified using by library barcodes to display different and common cell populations. Major differences between wild-type and mutant cell populations were marked by single or double asterisks respectively. (C) sox2 highlight progenitor cells while elavl3 mark developing neurons. (D) neurog1 and neruod1 identify neuronal progenitors and developing neurons. (E) Cells expressing dsRed and EGFP from transgenes introduced into shox2+/+ embryos. Cell clusters identified from graph-based clustering that overlapped with dsRed and EGFP in shox2+/+ and shox2Δ/Δ were used to identify progenitors and developing neurons. (F) Plot of progenitor and development obtained from identified subset of cell clusters were obtained for further analysis. Progenitor and developing neuron populations from shox2+/+ (magenta) and shox2Δ/Δ (green) cells were identified using library identification barcodes for each genotype. (G) Sub-population of cells express sox2 and elavl3 to mark progenitor and developing neurons, respectively. (H) Sub- population of cells express neurog1 and neruod1 to mark neuronal progenitors and developing neurons, respectively.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Biol. Open