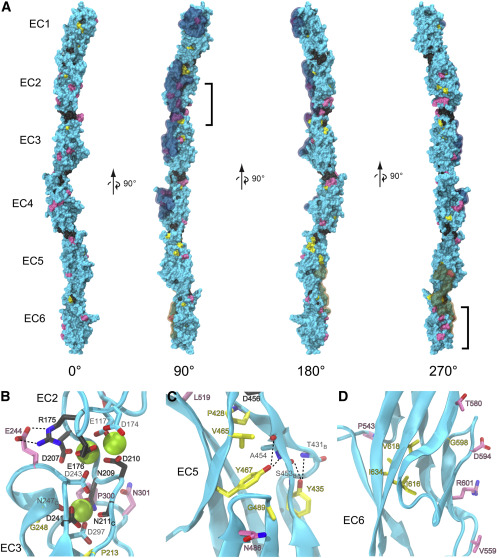
Fig. 6
Figure 6. PCDH19-CE mutations may affect Ca2+ binding, core packing, and surface properties of PCDH19 (A) Molecular surface representation of the composite dr Pcdh19 EC1-6 structure. Residues are colored based on the location of corresponding epilepsy mutations: Ca2+-binding motifs (dark gray), hydrophobic core (yellow), and surface (pink). The remaining residues without documented mutations in the literature are colored cyan. Brackets highlight two clusters of surface mutations in EC2 and EC6. Residues observed in the previously documented trans adhesion interface of dr Pcdh19 (Cooper et al., 2016) are highlighted in a semitransparent cyan. The cis interface only observed thus far in clustered protocadherins (Goodman et al., 2017) was mapped onto dr Pcdh19 and highlighted in semitransparent orange. (B–D) Ribbon diagrams highlight details of the EC2-3 linker region (B), EC5 hydrophobic core (C), and EC6 surface residues (D). Side chains are represented in stick for all mutant residues with carbon atoms matching the coloring of (A). Backbone atoms are also shown in stick for some residues. “B” subscripts indicate only the backbone is represented for clarity, and “C” subscripts indicate only the carboxyl group of the backbone is represented. Residue labels are colored according to the mutations category and outlined in black, while other relevant residues without documented mutation have cyan colored carbons and light gray labels. Numbering in the labels corresponds to the structure's numbering, and corresponding numbering of the human mutations can be found in Figure 4 and Data S4. Dashed lines depict hydrogen bonding discussed in the main text.
Reprinted from Structure (London, England : 1993), 29(10), Hudson, J.D., Tamilselvan, E., Sotomayor, M., Cooper, S.R., A complete Protocadherin-19 ectodomain model for evaluating epilepsy-causing mutations and potential protein interaction sites, 1128-1143.e4, Copyright (2021) with permission from Elsevier. Full text @ Structure