Fig. 4
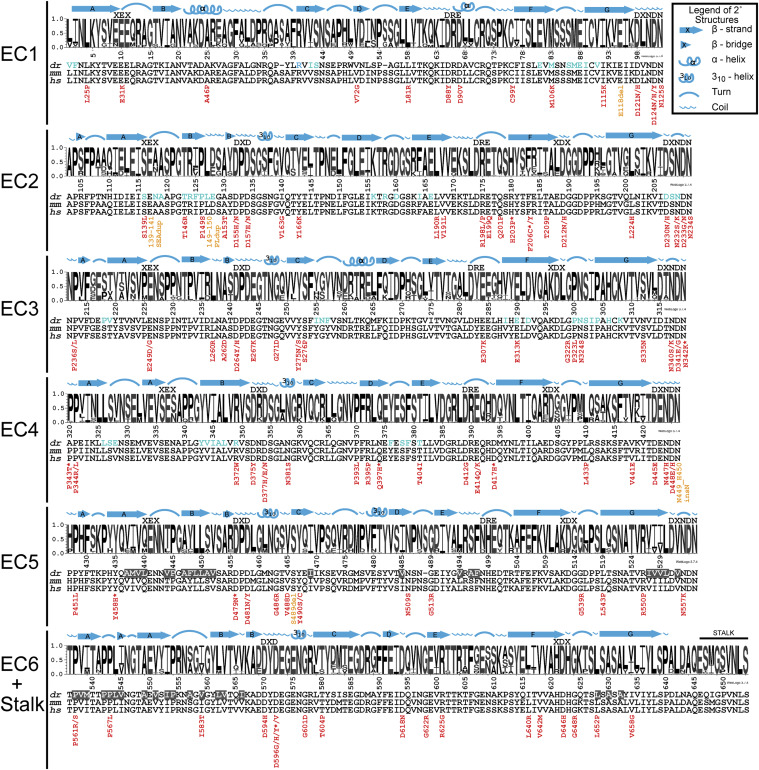
Figure 4. Ectodomain epilepsy mutations and structural features mapped onto sequence conservation Sequence logos represent the portion of species containing each amino acid by the height of that letter in the stack position (y axis units, probability). The numbering of the x axis represents the mature dr PCDH19 protein (signal peptide already cleaved off), which matches the numbering in the dr PCDH19 structure. Above each logo, the sequences of canonical cadherin repeat motifs are listed, and the secondary structural elements observed in the dr PCDH19 composite structure are drawn. For regions that do not have experimental data indicating secondary structures, features predicted from the protein sequence are labeled above a black line. Sequences for the most common model organisms used to study Pcdh19 (dr, Danio rerio; mm, Mus musculus) are aligned with the human sequence (hs, Homo sapiens). Residues found at the Pcdh19 trans interface (as presented in Cooper et al., 2016) are indicated with blue text in the dr sequence, and residues identified at the hydrophobic patch of dr Pcdh19 are in white text with gray background. Each of the epilepsy-causing mutations is listed below the human sequence with the missense mutations in red and in-frame insertions/duplications in orange. An asterisk denotes a patient with mutations affecting two or more PCDH19 amino acids. See also S5 and Figure S8.
Reprinted from Structure (London, England : 1993), 29(10), Hudson, J.D., Tamilselvan, E., Sotomayor, M., Cooper, S.R., A complete Protocadherin-19 ectodomain model for evaluating epilepsy-causing mutations and potential protein interaction sites, 1128-1143.e4, Copyright (2021) with permission from Elsevier. Full text @ Structure