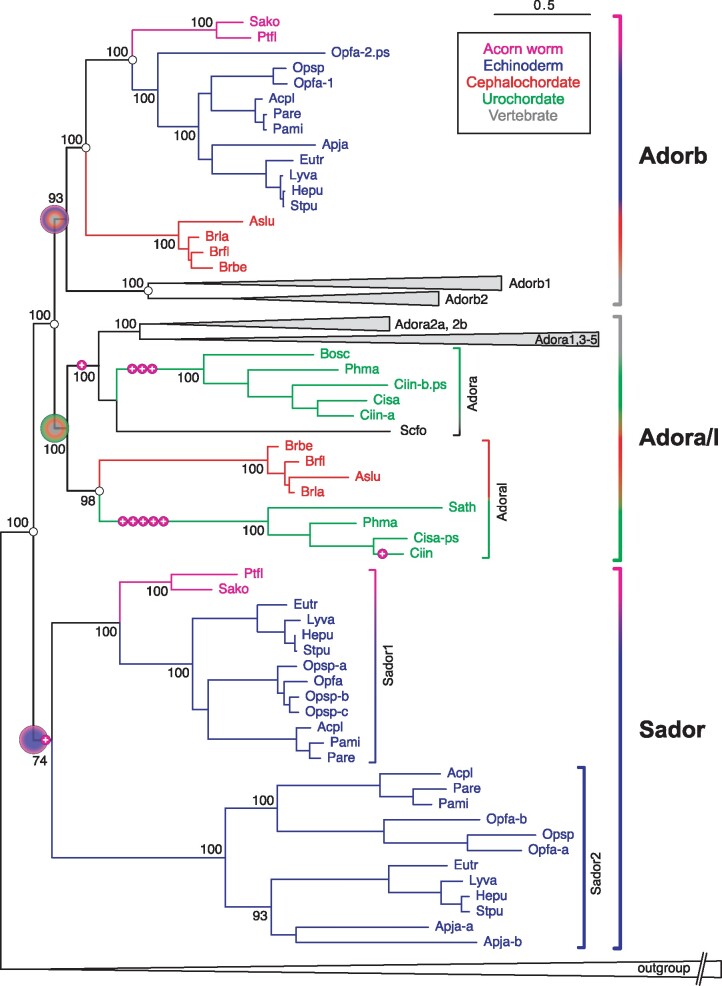
Fig. 1
The evolutionary origin of the nucleotide receptor adorb in the MRCA of deuterostomes. Maximum likelihood tree for adora, adorb, and related receptors. Branch support is given in %, scale bar denotes number of amino acid substitutions per site. Species groups are color-coded as indicated, all vertebrate clades are collapsed. Magenta circles with plus sign indicate intron gains, small empty circles denote absence of introns. Gene clades are visualized by angular parentheses, color-coded according to the species represented in that clade. Species are indicated by four letter abbreviation, for full names see supplementary file 3, Supplementary Material online. The root of the adora, adorb, and sador clade is denoted by large circles, color-coded according to species represented within that clade. Adora, adoral, adorb, and sador clades are well separated with high to maximum branch support. Adorb is present in all deuterostome groups analyzed except urochordates. The outgroup (collapsed) consists of over 100 genes encompassing all subgroups of rhodopsin class GPCRs as well as rhodopsin-associated class genes (see supplementary file 3, Supplementary Material online).