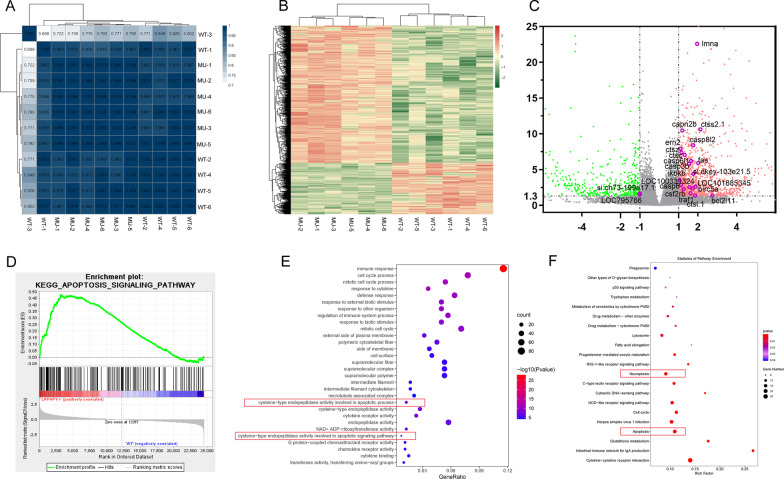
Fig. 4
Results of RNA sequencing and bioinformatics analysis. A Correlation matrix. The darker the blue, the higher the correlation coefficient. B Heat map showing differentially expressed genes in the eyes of lrpap1 mutant versus wild-type zebrafish. The log10 (TPM expression levels + 1) value is color-coded. C Volcano plot analysis of differentially expressed genes in lrpap1-deficient versus wild-type animals at three months post-fertilization. The abscissa represents the change in gene expression multiple in different samples, and the ordinate represents the statistical significance of the change of gene expression. The orange dots represent 91 genes significantly up-regulated in the mutant group, and the green dots represent 247 down-regulated genes. The purple circles show 22 genes associated with apoptosis factors. D GSEA analysis in the context of apoptosis pathway gene sets (NES = 1.642, p value = 0.010). E, F GO and KEGG analysis of the differentially expressed (|log2(Fold Change)|> 1 and Q value < 0.05). Apoptotic-related pathways are highlighted with red boxes. WT, wild-type. MU, lrpap1 homozygous mutant