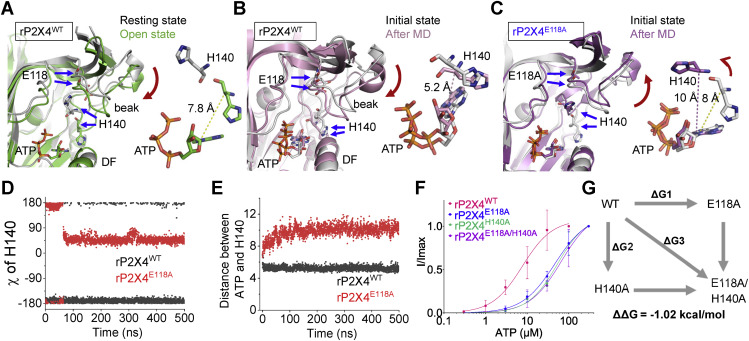
Fig. 7
Figure 7. H140 mediates the coupling of E118 to the ATP recognition of rP2X4. A, superimposed apo and open structures of rP2X4 to show the downward motions of the head domain and beak region, as well as the H140s moving closing to the bound ATP. Arrows highlight bound ATP–induced allostery. The yellow-dashed line indicates the distance between the Cα of H140 and N9 atom of ATP. B and C, superimposition of the initial open structure and averaged conformations of rP2X4WT (B) and rP2X4E118A (C) after 0.5-μs molecular dynamics (MD) simulations. Red arrows indicate the conformational changes during MD simulations. D and E, fluctuations of the dihedral angle of H140 (D) and distance between ATP and H140 (E) during 0.5-μs MD simulations of rP2X4WT and rP2X4E118A. The trajectories were sampled every 200 ps. F, the ATP concentration–response curves of rP2X4WT, rP2X4E118A, rP2X4H140A, and rP2X4E118A/H140A (mean ± SEM, n = 3–5, EC50 = 53.3 ± 9.1 and 55.1 ± 14.4 μM, nH = 1.24 ± 0.16 and 0.99 ± 0.15 for H140A and E118A/H140A, respectively). The data on rP2X4WT and rP2X4E118A are the same as those displayed for Figure 4B. G, double-mutant cycle analysis between E118 and H140A. rP2X4, rat (Rattus norvegicus) P2X4.