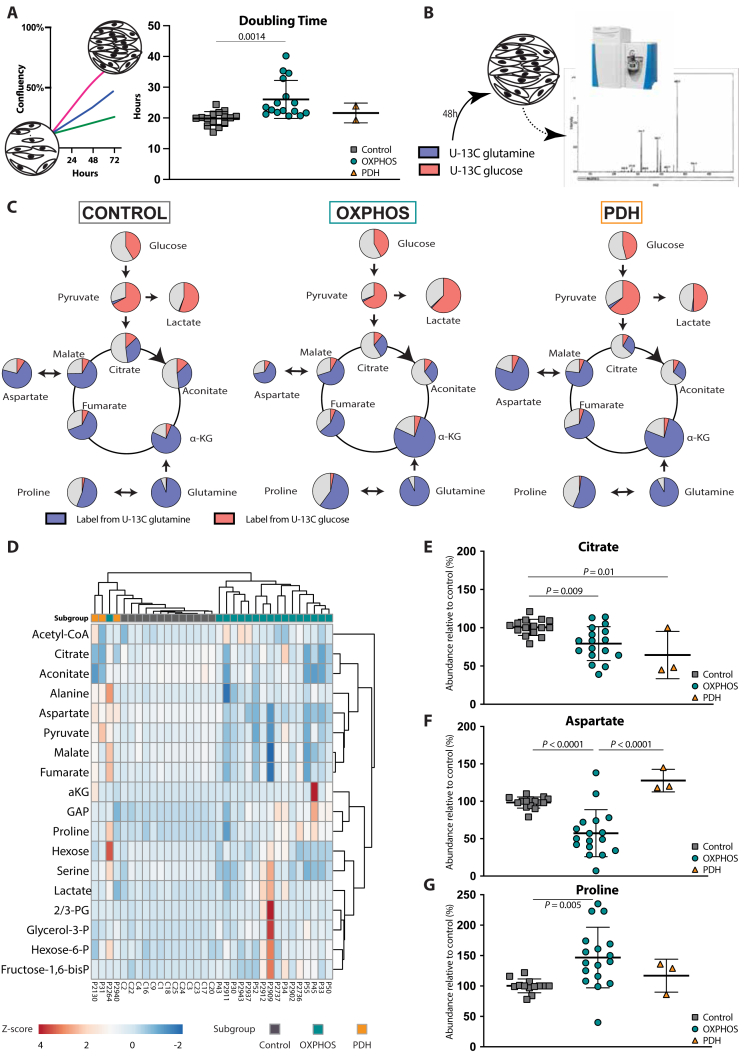
Fig. 1
Metabolic profile of mitochondrial disease A Design of the fibroblast proliferation assays based on 72-hours IncucyteⓇ proliferation experiments. Doubling time was calculated over a 48-hour period starting 24 h after plating (control (n = 15), OXPHOS deficient (n = 17) and PDH deficient (n = 3) fibroblast cell lines; technical replicates >3 per cell line). B Design of the tracer-based metabolomics studies with cultures with U–13C glucose or U–13C glutamine. C A simplified map of glycolysis and TCA cycle is depicted. The area of the circles represents the relative abundance of the metabolite compared to average control. Pink, blue and grey colour represent the fractional labelling coming from U–13C glucose, U–13C glutamine or other sources, respectively. D Heatmap displaying the changes in metabolite relative abundances of glycolysis, TCA and linked amino acids relative to controls in the different fibroblast cell lines. The colour scale (blue to red) corresponds to the z-score value of the relative abundance of the metabolite. Grouping of the different cell lines into disease subgroups is annotated with different colours:  OXPHOS deficiency;
OXPHOS deficiency;  PDH deficiency and
PDH deficiency and  controls. Full description of the genetic deficiency per cell line is described in Table 1. E The relative abundances of citrate, F aspartate and G proline in control, OXPHOS deficient and PDH deficient fibroblasts. Tracer metabolomics in control (n = 15), OXPHOS deficient (n = 17) and PDH deficient (n = 3) fibroblast cell lines (technical replicates 1–3 per cell line). Abundances normalized for protein content (BCA) and to the average of the controls per experiment. Statistics: one-way ANOVA with post-hoc Dunnett's T3 multiple comparison tests, and the error bars are +/−SD. αKG: alpha-ketoglutarate; CoA: coenzyme A; DHAP: dihydroxyacetone phosphate; Fructose-1,6-bisP: fructose-1,6-bisphosphate; GAP: glyceraldehyde-3-phosphate; Glycerol-3-P: Glycerol-3-phosphate; Hexose-6-P: Hexose-6-phosphate; OXPHOS: oxidative phosphorylation system; PDH: pyruvate dehydrogenase; PEP: phosphoenolpyruvate; TCA: tricarboxylic acid; 2/3-PG: 3-phosphoglycerate.
controls. Full description of the genetic deficiency per cell line is described in Table 1. E The relative abundances of citrate, F aspartate and G proline in control, OXPHOS deficient and PDH deficient fibroblasts. Tracer metabolomics in control (n = 15), OXPHOS deficient (n = 17) and PDH deficient (n = 3) fibroblast cell lines (technical replicates 1–3 per cell line). Abundances normalized for protein content (BCA) and to the average of the controls per experiment. Statistics: one-way ANOVA with post-hoc Dunnett's T3 multiple comparison tests, and the error bars are +/−SD. αKG: alpha-ketoglutarate; CoA: coenzyme A; DHAP: dihydroxyacetone phosphate; Fructose-1,6-bisP: fructose-1,6-bisphosphate; GAP: glyceraldehyde-3-phosphate; Glycerol-3-P: Glycerol-3-phosphate; Hexose-6-P: Hexose-6-phosphate; OXPHOS: oxidative phosphorylation system; PDH: pyruvate dehydrogenase; PEP: phosphoenolpyruvate; TCA: tricarboxylic acid; 2/3-PG: 3-phosphoglycerate.