Image
Figure Caption
Fig. 1
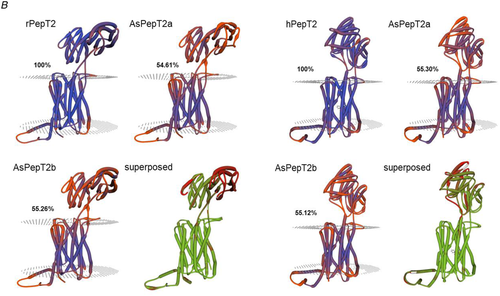
A, multiple alignments of the Atlantic salmon PepT2a (AsPepT2a) and PepT2b (AsPepT2b), zebrafish PepT2 (zfPepT2), rat PepT2 (rPepT2) and human (hPepT2) amino acid sequences as obtained by using ClustalX 2.1 and edited in GeneDoc 2.7 software. The conserved PTR2 family proton/oligopeptide symporter signature 1 (PROSITE pattern PS01022 – amino acid residues 100−124 on rPepT2 and hPepT2, and 86–110 on zPepT2, AsPepT2a and AsPepT2b) is marked by orange diamonds (♦). The putative transmembrane domains, named I to XII, were drawn using the annotation data of rPepT2 (see UniProtKB Acc. No. Q63424). Only one conserved extracellular N-glycosylation site (PROSITE pattern PS00001 – amino acid residues 528–532 on rPepT2 and hPepT2, 513–516 on zPepT2, and 511–514 on AsPepT2a and AsPepT2b), as obtained using NetNGlyc 1.0 server, is reported and marked by brown circles (●). Highlighted are key residues referring to the ‘extracellular gate’ (green triangles, ▲), ‘intracellular salt bridge’ (red triangles, ▲), and ‘peptide binding’ (blue triangles, ▲), as defined on the Cryo-EM structure of the rat PepT2 transporter (Protein Data Bank Acc. No. 7nqk.1; Parker et al., 2021). Key residues referring to the mechanism for substrate recognition and transport, as defined on the Cryo-EM structure of the human PepT2 (Protein Data Bank Acc. No. 7pmy.1) or human PepT1 (Protein Data Bank Acc. Nos. 7pn1.1, 7pmx.1 and 7pmw.1; Killer et al., 2021) are also highlighted. Amino acid residues involved in the subsequent steps of the transport cycle are shown: ‘outward-facing open’ state (apo) (1) (adapted from the human PepT1 structure, ref. Protein Data Bank Acc. No. 7pn1.1), ‘outward-facing open’ state (substrate-bound), (2) (adapted from the human PepT1 structure, ref. Protein Data Bank Acc. No. 7pmx1.1), ‘outward-facing occluded’ state (substrate-bound), (3) (adapted from the human PepT1 structure, ref. Protein Data Bank Acc. No. 7pmw1.1), ‘inward-facing partially occluded’ state (substrate-bound), (4) (adapted from the human PepT2 structure, ref. Protein Data Bank Acc. No. 7pmy1.1), ‘inward-facing open’ state (apo), (5) (from the human PepT1 Alphafold structure prediction, ref. Alphafold Acc. No. AF-P46059-F1; Jumper et al., 2021). The amino acid residues involved in ‘substrate recognition’ are all specifically indicated, marked by blue squares (■) (from the human PepT2 structure, ref. Protein Data Bank Acc. No. 7pmy1.1). Substrate: Ala-Phe. For details on transport dynamics, mechanism, states and conformations, please see Killer et al. (2021). B, three-dimensional representation of single and superposed structures of rPepT2, AsPepT2a and AsPepT2b (‘outward-facing open’ conformation) (left), and hPepT2, AsPepT2a and AsPepT2b (‘inward-facing partially occluded’ conformation) (substrate-bound; the substrate Ala-Phe is represented) (right), as obtained by using SWISS-MODEL tools. The models were built using as a template the rPepT2 (Protein Data Bank Acc. No. 7nqk.1) or the hPepT2 (Protein Data Bank Acc. No. 7pmy.1); then, the models were superposed by using the ‘Compare’ view tool. Colour schemes for single structures based on (model) confidence (it evaluates, for each residue of the model, the expected similarity to the native structure, thus representing an index of the ‘local quality’ of the residue): red, low confidence and blue, high confidence. Colour scheme for superposed structures based on consistency (it identifies local deviations of a protein structure from the ‘consensus’ established by all other structures selected for comparison): red, low consistency and green, high consistency. Percentage identities of Atlantic salmon PepT2 proteins vs. the rat (left) and human (right) PepT2 proteins are reported. [Colour figure can be viewed at wileyonlinelibrary.com]
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ J. Physiol.