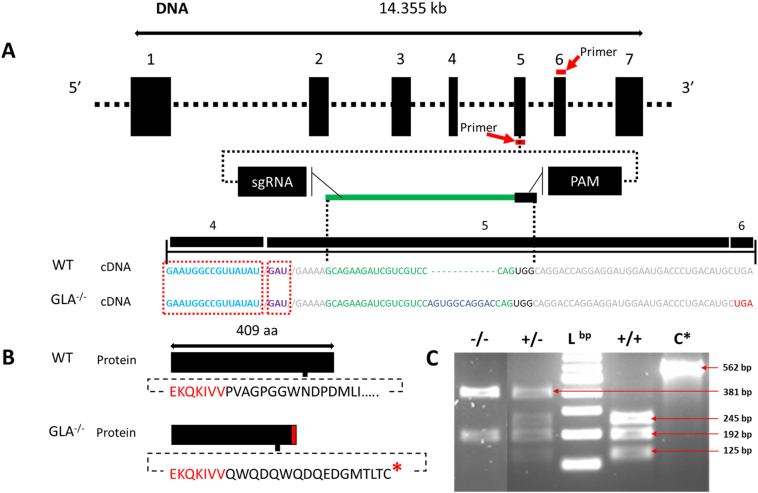
Fig. 3
Generation and verification of a GLA mutant line. (A) CRISPR/Cas9-based gene-editing tool used to generate mutant zebrafish. The targeted genomic sequence for the introduction of the insertion mutation in the exon 5 is illustrated. The eleven base pair insertion (shown in blue) is highlighted in the cDNA sequence. The putative stop codon after the insertion is highlighted in red. Guide RNA and PAM sequences are highlighted in green and black, respectively. Mutation occurred 45 bp from the proton donor catalytic site (in violet) and 120 bp from the substrate binding site (in light blue) that is in exon 4. Primers for genotyping and sequencing spanning exons 5 and 6 are indicated by the red bar (B) The insertion mutation leads to a frameshift with a premature stop codon (in red) 16 amino acid downstream the insertion region (the lower panel). (C) PCR screen shows that the insertion interrupts one of the two restriction enzyme digestion sites resulting in two PCR fragments in the mutant (−/−) instead of three in the wild type (+/+), Lbp = DNA ladder in base pairs (bp), C* = wild type undigested PCR product (the agarose gel image is edited for illustration, full image can be reviewed in sup.5). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)