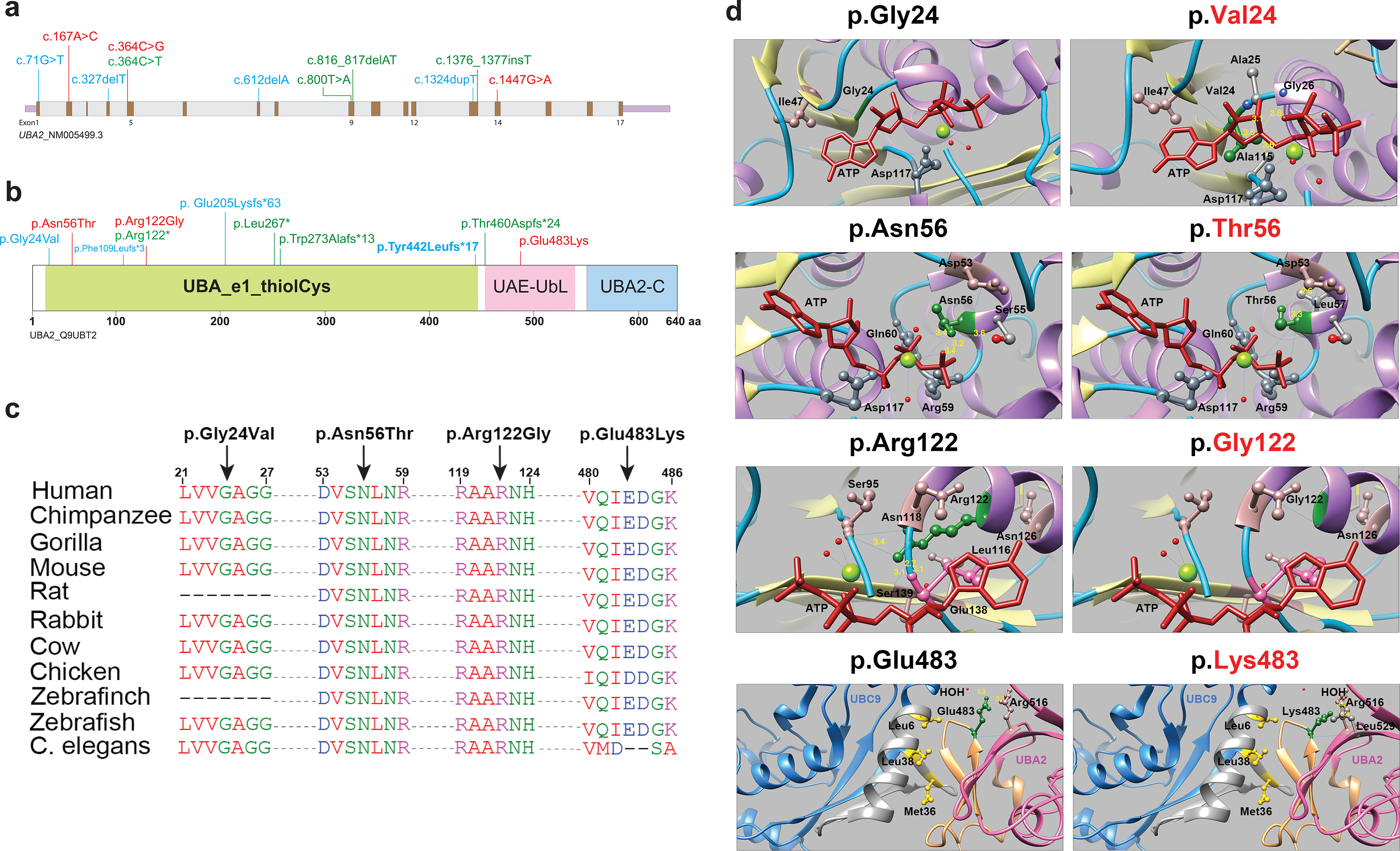
Figure 2: Fig. 2. UBA2 syndrome–associated variants and molecular modeling. (a) Schematic representation of the UBA2 gene. Exons are shown in brown color boxes; introns and 3’ and 5’ UTRs are in light gray and purple, respectively. Newly reported UBA2 missense and loss-of-function variants are shown in red and green, respectively, while blue is used to represent previously reported UBA2 variants. (b) Schematic representation of UBA2 protein domains. The UBA2 protein domain carrying catalytically active sites of ubiquitin-activating enzyme is shown in light green. This domain has putative active sites to bind ATP, substrate, and zinc with the last of five conserved cysteine residues playing an important role in ubiquitin thioester complex formation. UBA2-C (C-terminus) and UAE-UbL (ubiquitin-like) domains are shown in pink and blue, representing the C-terminus of UBA2 protein. UAE-UbL is structurally similar to ubiquitin and is involved in E1-SUMO-thioester transfer to E2 conjugation protein. The amino acid changes for the aforementioned variants are shown in the same color scheme as (b). (c) Amino acid sequence alignments of the human UBA2 protein across different species at each of the residues reported with missense variants. (d) Molecular modeling of human UBA2 protein. Secondary structure helix, strand and coil regions are shown in purple, yellow and cyan blue shades, respectively. Forest green color is used to show residues of interest in proteins with wild type (WT) and missense changes and the ATP molecule is shown in brick red color. Blue color shows regions of hydrogen bonding and light pink shows residues involved in hydrogen bond formation with residues of interest. The distances to nearby residues are shown by dashed yellow lines. Last panel: UBA2 and UBC9 are shown in hot pink and cyan blue color, respectively. As per molecular modeling predictions, p.Gly24Val: Glycine is flexible enough to maintain torsion angles and is buried in the protein core to maintain local secondary structure. p.Asn56Thr. Introducing a smaller but more hydrophobic residue at Asparagine 56 results in an empty space in the protein core and subsequent loss of hydrogen bonding with Asp53. p.Arg122Gly: The typical Arginine 122 residue is involved in hydrogen bonding with Asn118, Gly138 and Ser139. Replacement with Glycine is predicted to disrupt this array.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Genet. Med.