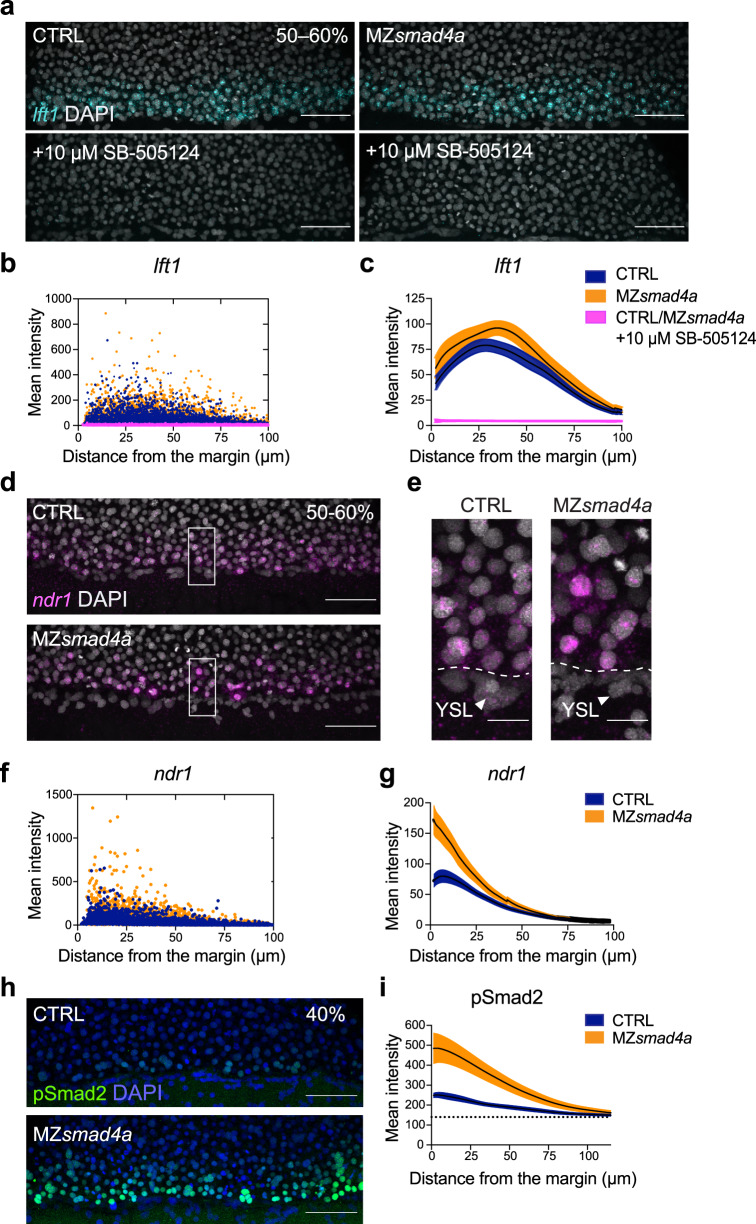
Fig. 4
a Maximum intensity projection (MIP) of quantitative ISH for lft1 at mid gastrulation (50–60% epiboly) in CTRL and MZsmad4a embryos treated ±10 µM SB-505124 at sphere stage. Scale bars correspond to 80 µm. Lateral views are shown. Nuclei were stained with DAPI (white). b Quantitation of a. Orange, dark blue, and magenta dots show segmented cells from CTRL and MZsmad4a embryos treated ±10 µM SB-505124. n = 8 embryos for each group. c Curve fitting for segmented cell intensities shown in b. Means are shown by black lines and the orange/dark blue shading indicates the SEMs. d, e MIP of quantitative ISH for ndr1 in CTRL and MZsmad4a embryos at mid-gastrulation (50–60% epiboly). Regions indicated in d by white boxes are shown enlarged in e. Scale bars correspond to 80 µm in d and 20 µm in e. Arrowheads indicate YSL. Lateral views are shown. Nuclei were stained with DAPI (white). f Quantitation of d. Orange and dark blue dots show segmented cells from CTRL and MZsmad4a embryos. n = 8 embryos for each group. g Curve fitting for segmented cell intensities shown in f. Means are shown by black lines and the orange/dark blue shading indicates the SEMs. h MIP of CTRL and MZsmad4a at 40% epiboly, immunostained for pSmad2. Scale bar corresponds to 80 µm. Lateral views are shown. Nuclei were stained with DAPI (blue). i Quantitation of the pSmad2 immunostaining at the margin of CTRL and MZsmad4a embryos. Each trace represents n = 8 embryos. Means are shown by black lines and the orange/dark blue shading indicates the SEMs. Dotted line shows baseline.