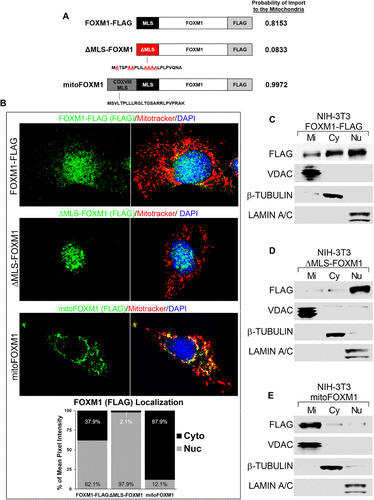
Fig. 2 Mitochondrial localization sequence in FOXM1 is required for translocation of FOXM1 to mitochondria. (A) Schematic representation of FLAG-tagged FOXM1 protein constructs with wild-type MLS (black box), mutated MLS (red box), and COX VIII MLS fused to FOXM1 (gray–black box). The probability of each construct to be imported to the mitochondria was determined using MitoProt (Claros and Vincens, 1996). (B) Cellular localization of FLAG-tagged FOXM1 fusion proteins. NIH-3T3 cells transfected with FOXM1-FLAG, ΔMLS-FOXM1, or mitoFOXM1 were used for colocalization studies with FLAG antibody (exogenous FOXM1) (green) and MitoTracker dye (red). Nuclei were counterstained with DAPI. Magnification: ×100; 20 μm scale bar. The intensity of FOXM1 fluorescence signal (green) in cytoplasmic (Cyto) and nuclear (Nuc) compartments was determined using CellProfiler software and is presented as the percent of mean pixel intensity (n = 7/group). (C–E) Western blots show the efficient localization of FOXM1 FLAG-tagged protein into different cellular compartments. NIH-3T3 cells expressing FOXM1-FLAG (C), ΔMLS-FOXM1 (D), or mitoFOXM1 (E) constructs were used to prepare mitochondrial (Mi), cytoplasmic (Cy), and nucleic (Nu) fractions. Western blots were probed with antibodies against FLAG (exogenous FOXM1), VDAC (mitochondrial protein), β-TUBULIN (cytoplasmic protein), and LAMIN A/C (nuclear protein).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Mol. Biol. Cell