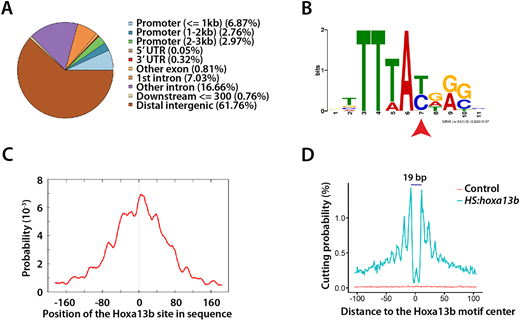
Fig. 3 Hoxa13b-binding sites and motif analysis. (A) Distribution of the Hoxa13b target sites in the genome. Most sites are in distal intergenic regions. (B) The Hoxa13b-binding motif discovered from analysis of the CUT&RUN data. The red arrowhead indicates a base that, as a T, is bound by all Hox proteins, whereas, as a C, it is bound by posterior Hox proteins, based on in vitro studies. (C) The Hoxa13b motif shown in B is centrally located among all the CUT&RUN DNA fragments that contain a Hox motif, as expected. (D) Hoxa13b forms a 19 bp footprint at the Hoxa13b motif sites in the HS:hoxa13b-FLAG-GFP sample (teal), whereas, in the control sample, DNA cutting by MNase was random and rare at the same sites (red).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development