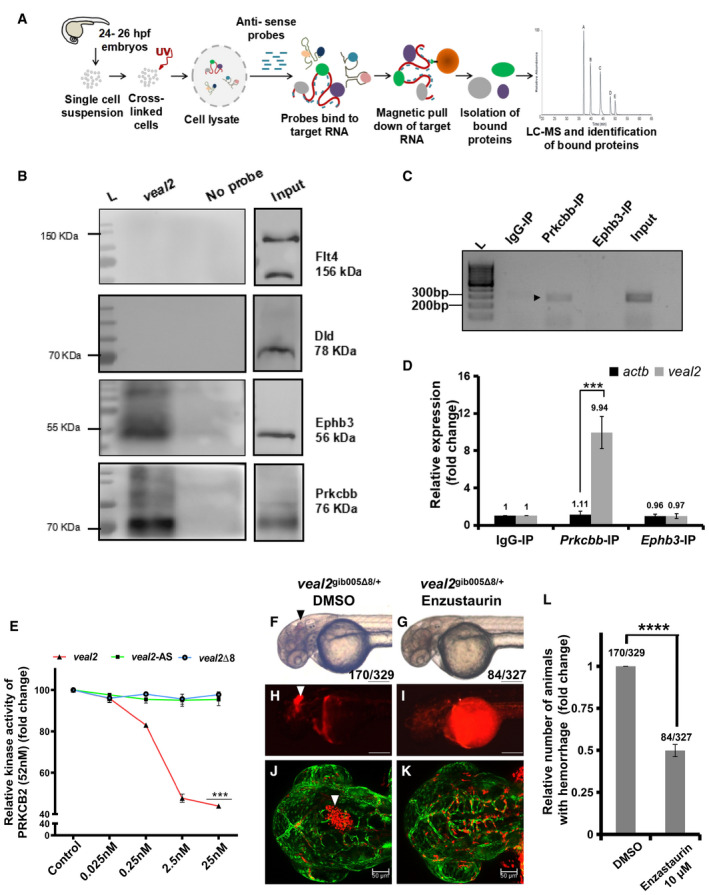
Fig. 3
Schematic of the methodology adopted for the identification of protein interacting partners of RAP‐MS followed by Western blotting validated Prkcbb and Ephb3 as interacting protein partners with RNA immunoprecipitation of qRT–PCR‐based quantification of Relative kinase activity of human PRKCB2 under standard conditions and in the presence of various concentrations of the WT Enzastaurin treatment rescues hemorrhage phenotype in Relative number of animals that displayed the hemorrhage phenotype across the progeny of the outcross of
Data information: All the experiments