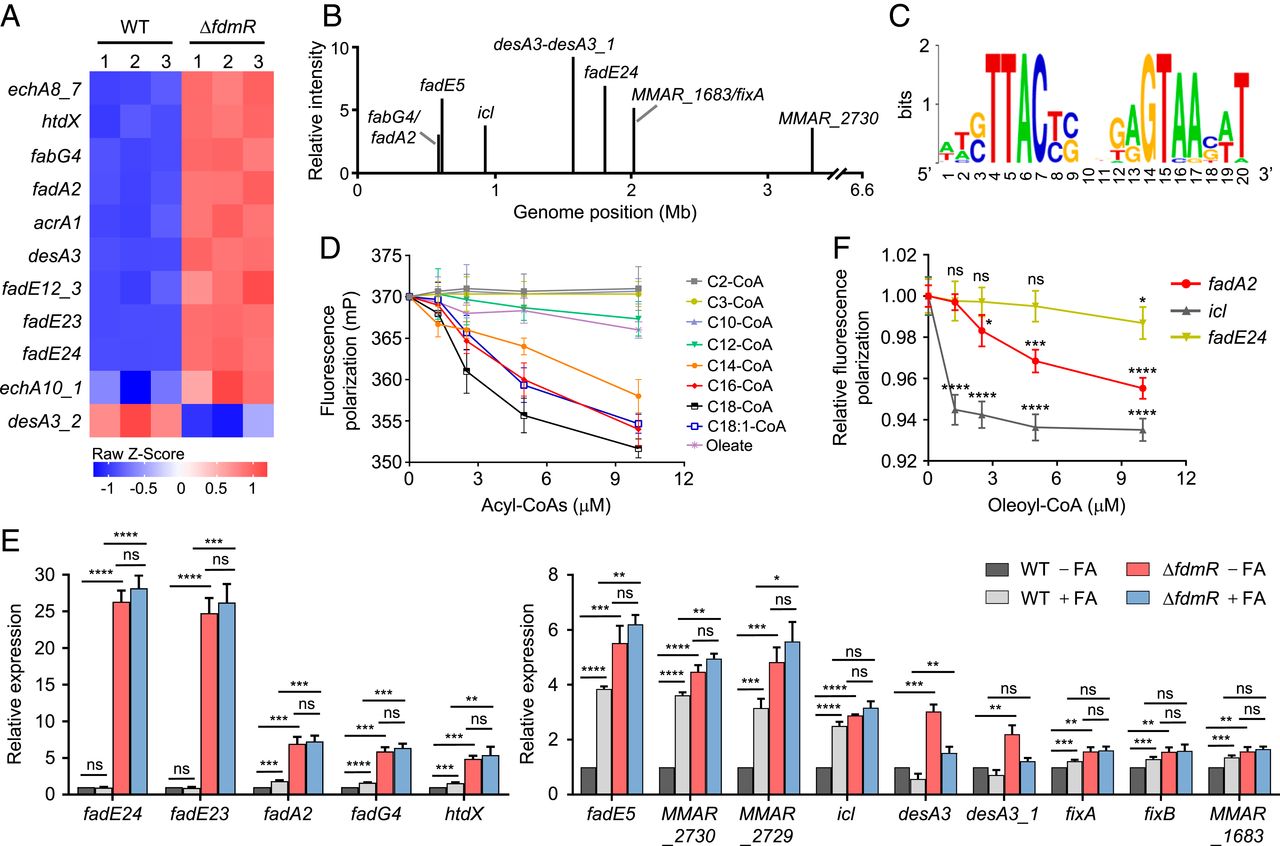
Fig. 2 Identification of FdmR regulon and effector molecules. (A) Heat map showing relative expression levels of the genes involved in fatty acid metabolism in M. marinum. The strains were grown in 7H9 medium with glycerol and OADC enrichment. Data shown are from three independent experiments. (B) Genome-wide FdmR-binding sites identified by ChIP-Seq. (C) Sequence logo of the FdmR-binding motif derived from ChIP-Seq enriched sequences. (D) Effect of various acyl-CoAs and oleate on the FdmR-DNA interaction determined by FPAs. The DNA operator of fadA2 was used. (E) Effect of long-chain fatty acid supplementation on expression of FdmR target genes in wild type and ∆fdmR mutant. The strains were grown in 7H9 with glycerol and glucose. Long-chain fatty acids were added to the medium as indicated. The expression levels of each gene were determined by qRT-PCR and normalized to the gene expression in the wild type grown in the absence of fatty acids. (F) Effect of oleoyl-CoA on the interaction of FdmR with various DNA operators. Data are normalized to the value in the absence of oleoyl-CoA. Data shown in D, E, and F are mean ± SD (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, not significant (ns), by unpaired two-tailed t test with false discovery rate correction.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Proc. Natl. Acad. Sci. USA