Fig. S3
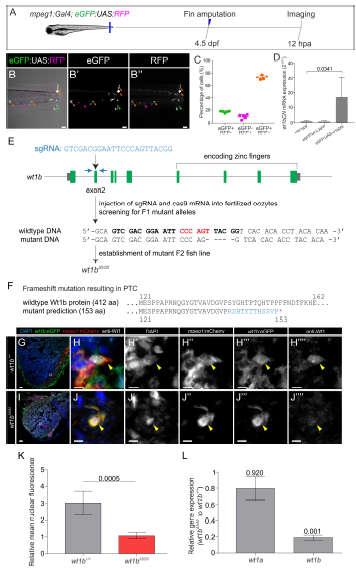
Validation of genetic lines to assess wt1b function. Related to Figures 3, 4 and 5. A-D, Validation of transgenic lines for macrophage-specific inhibition of wt1b function. A, Caudal fins of Tg(mpeg1:Gal4; GFP:UAS:RFP) larvae were amputated at 4.5 dpf and eGFP and RFP positive cells imaged at 12 hpa. B–B’’’, Images of the caudal fin showing RFP and eGFP positive cells. B’ and B’’ are single channels for eGFP and RFP. Orange arrowheads, double positive cells; magenta arrowheads, RFP+ cells; and green arrowheads, eGFP+ cells. C, Quantification of the percentage of eGFP+, RFP+ and double-positive cells. Dots indicate individual fish, shown are means Å} SD. Note that over 70% of macrophages are double-positive. D, Confirmation of wt1bDN overexpression using the Tg(eGFP:UAS:wt1bDN) line. Gal4FF mRNA was injected into 1-cell stage wildtype, embryos from the transgenic control line Tg(eGFP:UAS:RFP) or Tg(eGFP:UAS:wt1bDN). qRT-PCR was performed on cDNA obtained from these larvae. Shown are mean values Å} SD (n= 4 biological replicates per condition). Statistical analysis by Mann-Whitney test. E-L , Generation of the wt1bΔ5 mutant line. Related to Figures 4 and 5. E , Schematic drawing of the wt1b gene locus and workflow for wt1b mutant generation. A fish line that lacks 5 nucleotides in exon 2 of the wt1b gene (wt1bΔ5 ) was established. Blue arrows indicate the forward and reverse primer positions and the BsrI restriction site, used to detect indels, is highlighted in red. The sgRNA target sequence is shown in bold. F , The 5 nucleotides deletion is predicted to result in a frame shift (blue) and a premature termination codon that leads to a truncation of the protein. G-J’’’’ , Immunostaining with anti-WT1, anti-GFP and anti-mCherry on cryoinjured heart sections at 4 days postinjury from wt1b+/+ ; wt1b :eGFP;mpeg1 :mCherry and wt1bΔ5/Δ5 ; wt1b :eGFP;mpeg1 :mCherry zebrafish. Cell nuclei are counterstained with DAPI. Colocalization (arrowheads) of Wt1 with GFP and mCherry is detected in wt1b+/+ but absent in wt1bΔ5/Δ5 . K , Quantification of the relative mean Wt1 nuclear fluorescence in images of wt1b :eGFP; mpeg1 :mCherry cells normalized by background signal. Two-tailed unpaired t test. L , Quantitative RT-qPCR analysis of wt1a and wt1b expression in wt1bΔ5/Δ5 vs. wildtype larvae. Error bars represent standard error. p-values are calculated by pair wise fixed reallocation randomization with REST (n=10). dpf, days post fertilization; hpa, hours post amputation. Scale bars 50 μm (G and I), 40 μm (B-B’’) and 5 μm (H-H’’’’ and J-J’’’’).