Fig. 7
Reciprocal Interactions of Fast and Slow Muscle Fibers Guides Their Differentiation
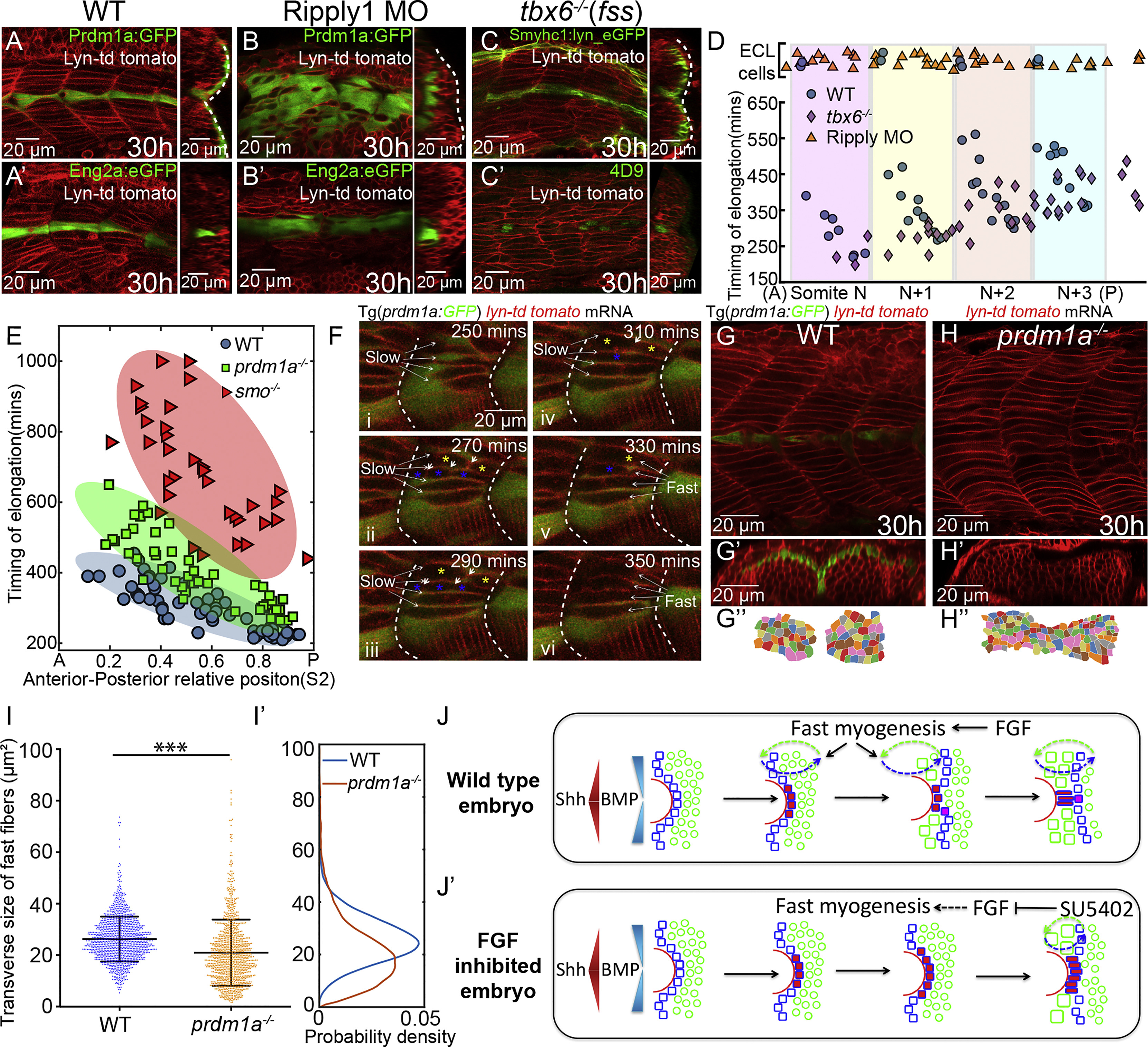
(A–C) The lateral migration of SSFs (A–C) and MPs specification (Aʹ–Cʹ) in wild-type embryos (A), Ripply1 morphants (B), and tbx6−/− mutants (C). Slow muscles are either labeled with Prdm1a:GFP (A and B) or Smyhc1:lyn_eGFP (C). MPs are either labeled with Eng2a:eGFP (Aʹ and Bʹ) or 4D9 antibody (Cʹ). Images are taken at muscle segments 12–16 at 30 hpf.
(D) The timing of elongation of individual fast muscle progenitors along the DV midline from a region corresponding to four adjacent somites. Shaded regions represent individual somites in wild-type embryos. Blue, orange, and violet dots denote cells from wild-type embryos (nEmbryos = 5), Ripply1 morphants (nEmbryos = 5), and tbx6−/−mutants (nEmbryos=5), respectively. Cells with ECL cell fate are placed at the top of the panel.
(E) AP dependence of the timing of elongation relative to somite segmentation of individual fast muscle progenitors along the DV midline in wild-type embryos (nCells = 57 from total of 7 somites taken from 6 embryos), prdm1a−/− (nCells = 70 from total of 6 somites taken from 6 different embryos), and smo−/− mutants (nCells = 42, from total of 4 somites taken from 4 different embryos). The ellipse represents the minimum-volume covering of data points in each group.
(F) Time-lapse of the patterning of fast muscle fibers in wild-type embryos at muscle segments 16–18. The fusing fast muscle progenitors are labeled with blue and yellow asterisks. White short arrows label the dissolving membrane between the fusing cells. The intercalated slow and fast muscles are labeled with long white arrows.
(G–H) Parasagittal optical view of wild-type embryos and prdm1a−/− mutant labeled with membrane marker lyn-td tomato at muscle segments 12–15. Transverse views of (G) and (H) are displayed in (Gʹ) and (Hʹ) with dorsal to the left and lateral to the top and manually segmented in (Gʹʹ) and (Hʹʹ).
(I) Distribution of the transverse size of fast muscle fibers in wild-type embryo (nCells = 1246 from total of 15 muscle segments taken from 8 embryos) and prdm1a−/− mutant (nCells = 1126 from total of 12 muscle segments taken from 6 embryos) (∗∗∗p < 0.001, Student’s t test). (Iʹ) Probability density of (I).
(J) Schematic presentation of how FGF signaling regulates MPs differentiation in wild-type embryos by precisely controlling the timing of SSFs migration and hence their exposure to Shh and BMP signaling. (Jʹ) In the absence of FGF signaling, ectopic MPs are induced not only at the expense of eng+ SSFs but also in an extended time window due to the delay of lateral migration.
Reprinted from Developmental Cell, 46, Yin, J., Lee, R., Ono, Y., Ingham, P.W., Saunders, T.E., Spatiotemporal Coordination of FGF and Shh Signaling Underlies the Specification of Myoblasts in the Zebrafish Embryo, 735-750.e4, Copyright (2018) with permission from Elsevier. Full text @ Dev. Cell