Fig. 1
Profiling of H3.3 Occupancy in Adult Zebrafish CMs
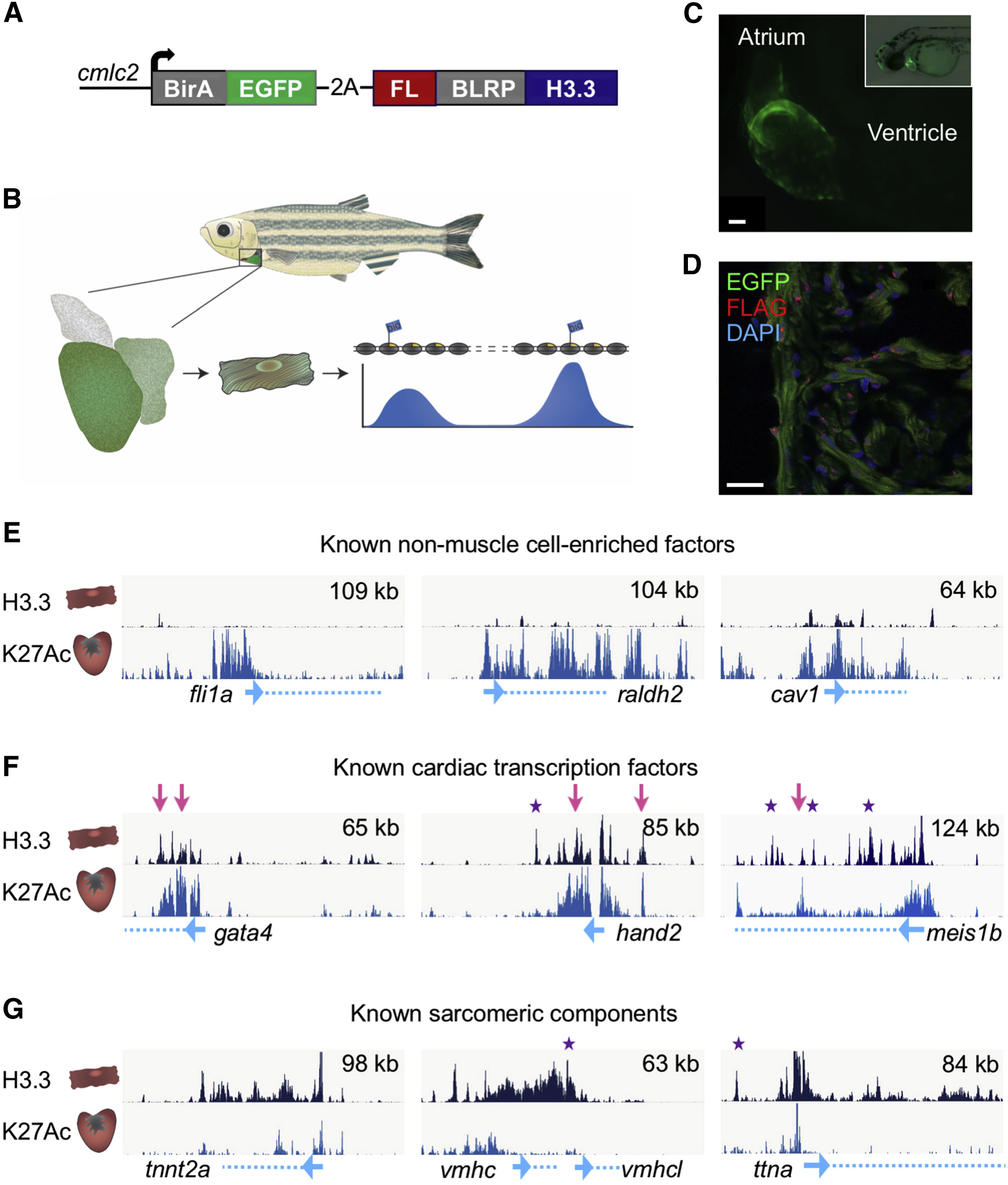
(A) The structure of the cmlc2:H3.3-bio transgene. EGFP is fused to a biotin ligase (BirA) followed by an in-frame 2A peptide sequence for polycistronic expression with the zebrafish histone H3.3 gene H3f3d. Fused in-frame to H3.3 are sequences coding an N-terminal FLAG and biotin ligase recognition peptide (BLRP).
(B) Schematic of H3.3-bio profiling.
(C) cmlc2:H3.3-bio larva 2 days post fertilization, showing cardiac EGFP expression. Scale bar, 20 μm.
(D) CMs (green) from adult uninjured cmlc2:H3.3-bio hearts contain nuclear H3.3-bio, detected with an anti-FLAG antibody (red). Scale bar, 20 μm.
(E) Genome browser snapshots indicating sparse H3.3 enrichment in and around genes with little or no expression in CMs (top track, dark blue). H3K27Ac from whole hearts (bottom track, light blue) is enriched around all promoters (blue arrows) and some gene bodies (dashed blue lines). y axis = enrichment of H3.3 = (Nc + 1)/(Nc)/(Ni + 1)/(Ni), where Nc = H3.3 ChIP, Ni = Input (top left). Top right, x axis (kb).
(F) Cardiac transcription factors are enriched with H3.3 throughout the gene body and in nearby intergenic regions. Many H3.3 peaks overlap with enrichment of H3K27Ac (pink arrows). Many other peaks are only enriched for H3.3 (purple stars).
(G) Sarcomeric protein genes are enriched for both H3.3 and H3K27Ac; however, there are many more H3.3 occupancy peaks.
Reprinted from Developmental Cell, 40, Goldman, J.A., Kuzu, G., Lee, N., Karasik, J., Gemberling, M., Foglia, M.J., Karra, R., Dickson, A.L., Sun, F., Tolstorukov, M.Y., Poss, K.D., Resolving Heart Regeneration by Replacement Histone Profiling, 392-404.e5, Copyright (2017) with permission from Elsevier. Full text @ Dev. Cell