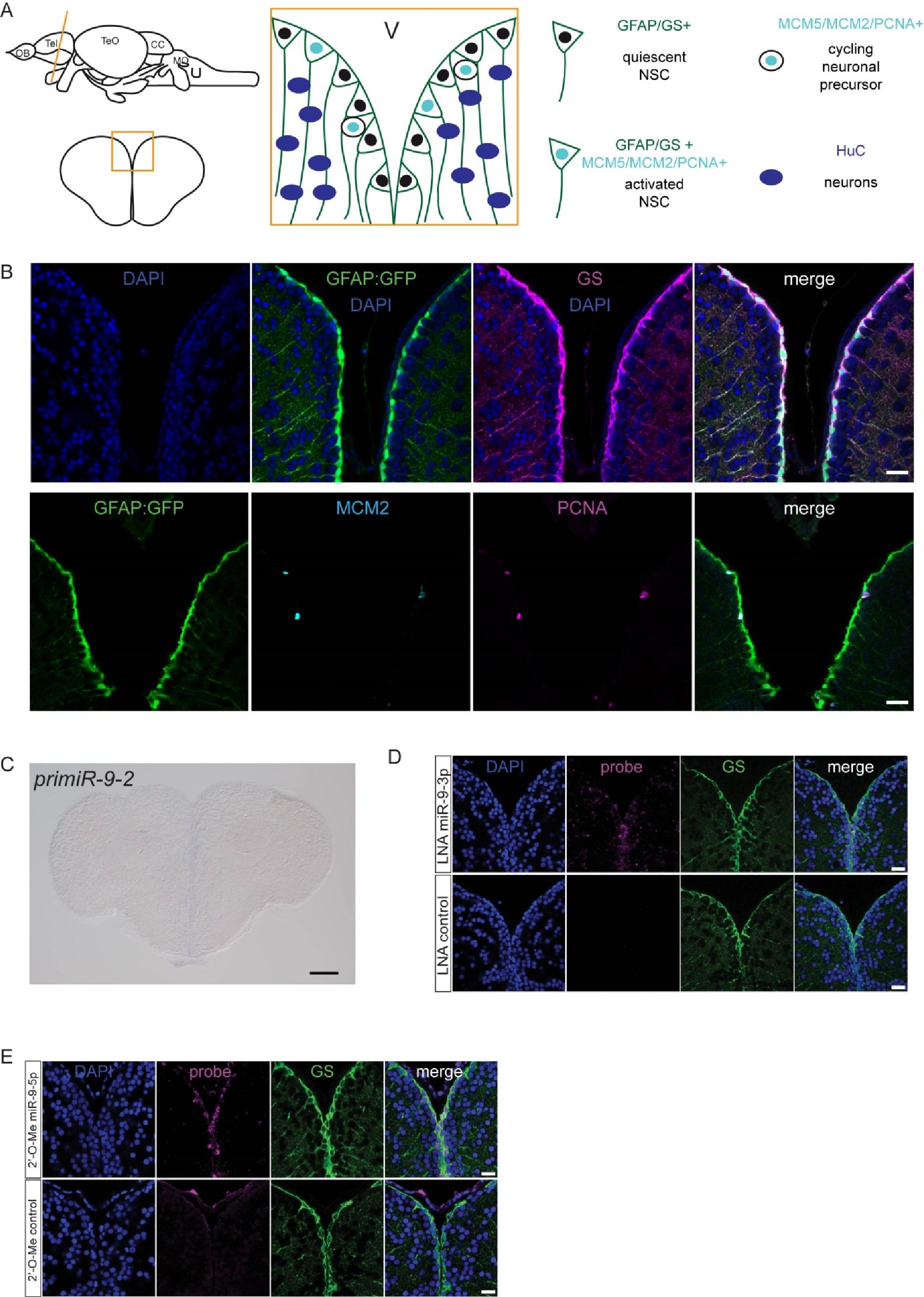
Fig. S1
Validation of markers and of the specificity of the LNA miR-9 probe, related to figure 1.
(A) Schematic representation of the germinal zone of the adult zebrafish pallium. Radial glia cells (GFAP+/GS+) lining the ventricle are bona fide NSCs and can be found in two states: activated or quiescent. The activated state is defined by the expression of proliferation markers (MCM2/MCM5/PCNA). Once activated, NSCs give rise to non-glial intermediate precursors which are committed towards neuronal differentiation. (B) Validation of the markers used in this study. All radial glia NSCs co-express GFP driven by the gfap promoter (green, gfap:GFP) and GS (magenta) (top panel). A small fraction of NSCs are found in an activated state in which they coexpress proliferation markers (MCM2, light blue; PCNA, magenta) (bottom panel). Scale bars=50μm (C) ISH of pri-miR-9-2 demonstrating that it is not expressed in the adult telencephalon. Scale Scale bar=100μm. (D) ISH using LNA probes for miR-9-3p (magenta, top panel) and a control sequence (magenta, bottom panel) with immunofluorescence for GS (green) and nuclear DAPI counterstaining (dark blue) demonstrating the specificity of the LNA probes used for mature miR-9-5p detection. In contrast to miR-9- 5p, miR-9-3p expression is much fainter and more diffuse although it is present at very low levels in the nucleus of some ventricular NSCs. (E) ISH using LNA 2'-O-Me probes for miR-9-5p (magenta, top panel) and a control sequence (magenta, bottom panel) with immunofluorescence for GS (green) and nuclear DAPI counterstaining (dark blue) confirming the exclusive expression of miR-9-5p in the ventricular NSCs. Scale bars=20μm.