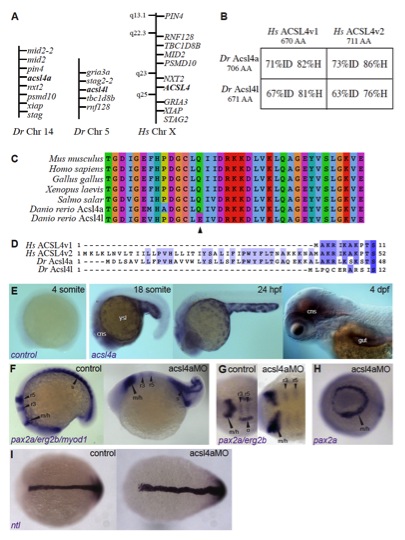
Fig. S1
Identification and embryonic expression of zebrafish acsl4a, which is necessary for proper dorsoventral patterning, related to Figure 1.
(A-D) Zebrafish Acsl4a is closely related to characterized ACSL4 proteins.
(A) Regions of conserved synteny demonstrate the relationship between human and zebrafish acsl4 orthologs. Syntenic analysis was performed using reciprocal BLASTN searches of the latest zebrafish (Ensembl Zv9) and human (Ensembl GRCh37) genomic assembly. These data were used to identify the genes adjacent to zebrafish acsl4 homologs and search for their human ortholog’s chromosomal locations. Genes surrounding both human ACSL4 and zebrafish acsl4 genes are shown.
(B) Amino acid sequence comparison of zebrafish (Danio rerio) Acsl4 proteins (Dr Acsl4a and Dr Acsl4l) to human ACSL4 splice isoforms (Hs ACSL4v1 and Hs ACSL4v2). Percentages shown are percent identity (ID) and homology (H) determined using BLOSUM62 matrix (Henikoff and Henikoff, 1992).
(C) Motif-II (Black et al., 1992; Kameda and Imai, 1985; Steinberg et al., 2000; Watkins et al., 2007) of ACSL4 homologs. Multiple sequence alignment of ACSL4 orthologs performed using the Jalview software (Clamp et al., 2004; Waterhouse et al., 2009). The MUSCLE algorithm was used with default settings (BLOSUM62). Acsl4l has an amino acid change from Glutamine to Glutamate at position 526 (arrowhead), an amino acid important for arachidonic acid specificity (Stinnett et al., 2007).
(D) Acsl4a contains the homologous N-terminal region of the longer human ACSL4 isoform. Multiple sequence alignment of zebrafish Acsl4 proteins and human ACSL4 splice isoforms performed using the Jalview with MUSCLE. Sequences are colored for percent identity.
(E) acsl4a expression in late stage embryonic development. Whole-mount mRNA in situ hybridizations using acsl4a sense control riboprobe (control) and antisense riboprobe (acsl4a). Lateral views, anterior to the left. ysl: yolk syncytial layer, cns: central nervous system.
(F-G) Whole-mount mRNA in situ hybridization of pax2a/erg2b/myod1 in 12-somite stage embryos either uninjected (control) or injected with 500 fmol acsl4a MO. pax2a marks m/h: midbrain/hindbrain boundary, o: otic vesicle. erg2b marks r3: rhombomere 3, r5: rhombomere 5. myod1 marks s: somites. (F) Lateral view, anterior to the left. (G) Dorsal view, anterior to the left.
(H) Whole-mount mRNA in situ hybridization of pax2a in 12-somite stage embryos either uninjected (control) or injected with 500 fmol acsl4a MO. Anterior view, dorsal to the right.
(I) Whole-mount mRNA in situ hybridization of ntl in 12-somite stage embryos either uninjected (control) or injected with 500 fmol acsl4a MO. Dorsal view, anterior to the left.
Reprinted from Developmental Cell, 27(6), Miyares, R.L., Stein, C., Renisch, B., Anderson, J.L., Hammerschmidt, M., and Farber, S.A., Long-Chain Acyl-CoA Synthetase 4A Regulates Smad Activity and Dorsoventral Patterning in the Zebrafish Embryo, 635-647, Copyright (2013) with permission from Elsevier. Full text @ Dev. Cell