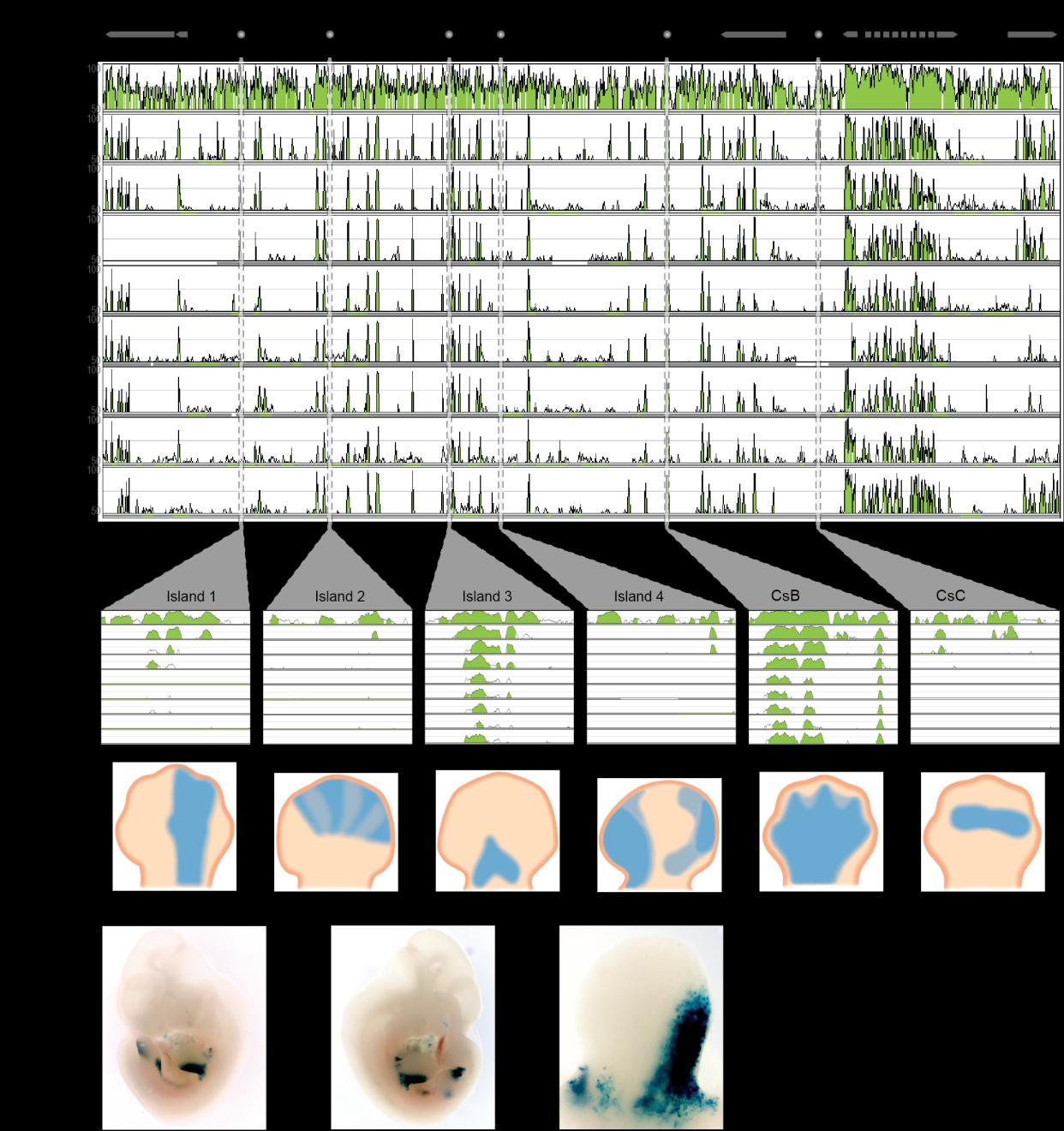
Fig. S14
Alignment of the HoxD locus and upstream gene desert identifies conserved limb enhancers. (a) Organization of the mouse HoxD locus and centromeric gene desert, flanked by the ATF2 and MTX2 genes. Limb regulatory sequences (I1, I2, I3, I4, CsB and CsC) are noted. Using the mouse locus as a reference (NCBI37/mm9 assembly), corresponding sequences from human, chicken, frog, coelacanth, pufferfish, medaka, stickleback, zebrafish and elephant shark were aligned. Alignment (mVISTA program, homology threshold 70%) shows regions of homology between tetrapod, coelacanth and ray-finned fishes. (b) Alignment of vertebrate cis-regulatory elements I1, I2, I3, I4, CsB and CsC. (c) Expression patterns driven by each regulatory element assayed via mouse transgenesis. (d) Expression patterns of coelacanth Island I in a transgenic mouse. Limb buds indicated by arrowheads in the first two panels. The third panel shows a close-up of a limb bud.