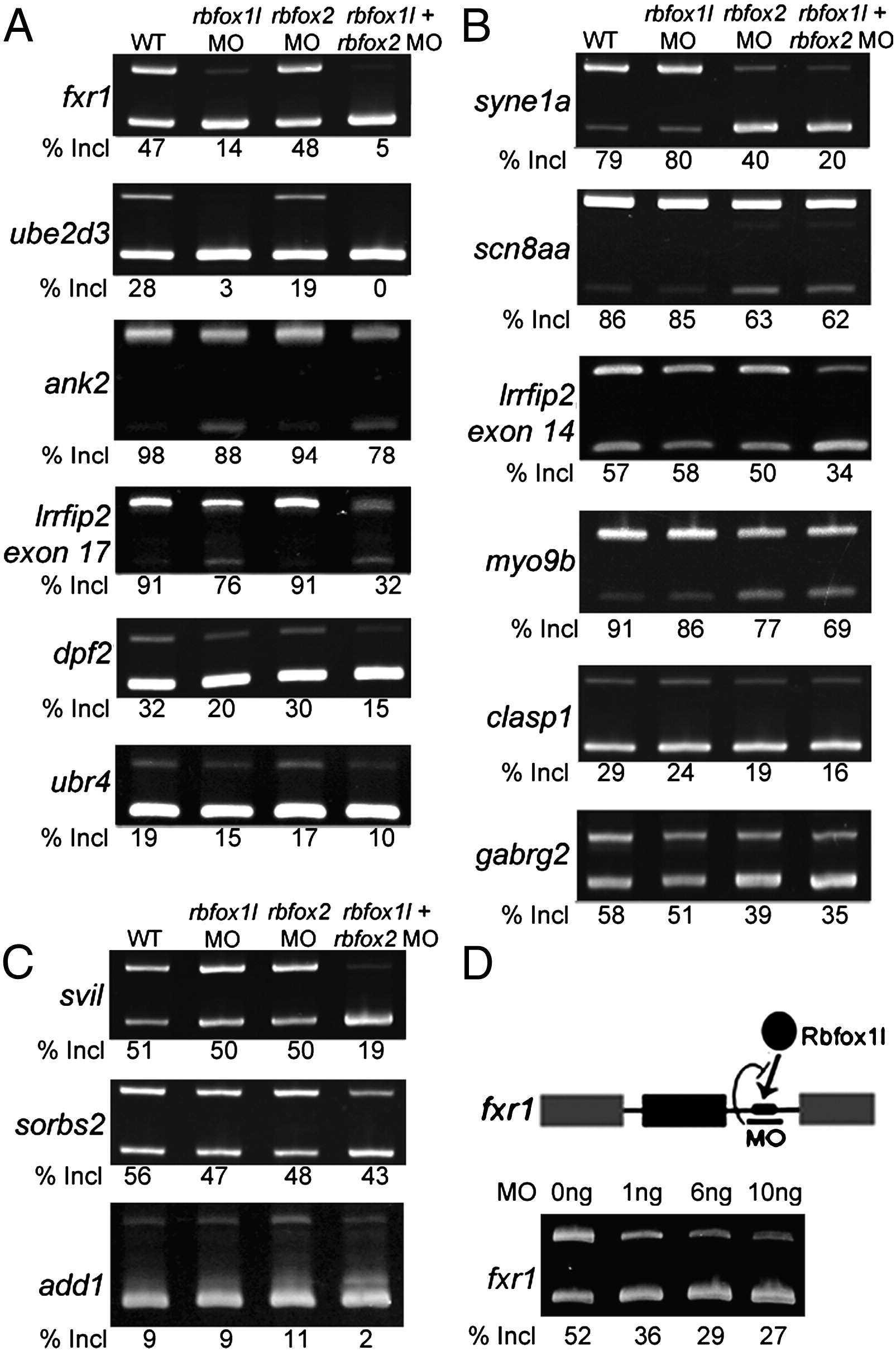
Fig. 3 Rbfox knockdown reveals three classes of regulated exons. Semi-quantitative RT-PCR splicing analysis of candidate target exons revealed exon skipping upon knockdown of rbfox1l (A), rbfox2 (B), or both rbfox genes (C). (D) An MO targeting the single Rbfox binding motif in the fxr1 intron reduced exon inclusion to a similar extent as knockdown of rbfox1l expression (A). All results were reproducible in at least 3 experiments (average exon inclusion levels noted below each gel). Alternative exon (black), constitutive exons (gray), Rbfox1l protein (black oval), Rbfox UGCAUG binding motif (black rectangle). In all panels, WT = mock-injected embryos except in D, WT = uninjected embryos, MO = morpholino, E = exon, Incl = inclusion. PCR primers and expected band sizes for inclusion and exclusion isoforms are listed in Supplementary Table S1.
Reprinted from Developmental Biology, 359(2), Gallagher, T.L., Arribere, J.A., Geurts, P.A., Exner, C.R., McDonald, K.L., Dill, K.K., Marr, H.L., Adkar, S.S., Garnett, A.T., Amacher, S.L., and Conboy, J.G., Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions, 251-61, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.