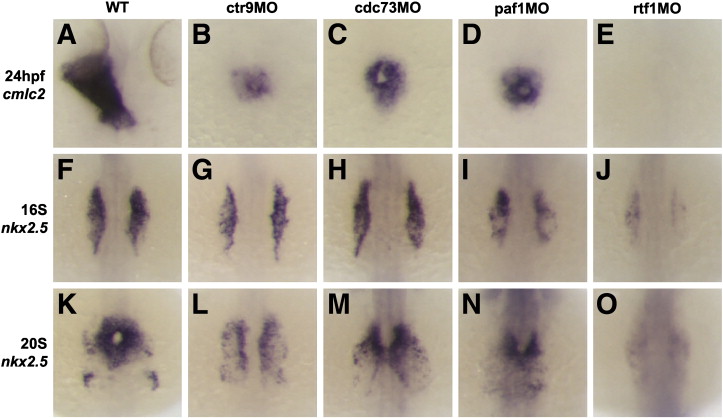
Fig. 3
Fig. 3. Loss of Ctr9, Cdc73, Paf1, and Rtf1 causes defects in myocardial migration, cardiomyocyte cell number, and primitive heart tube elongation. (A–E) Dorsal views of cmlc2 expression in 24 hpf wild type and morphant embryos. Wild type embryos have an elongated primitive heart tube at 24 hpf (A), whereas ctr9 (B), cdc73 (C), and paf1 (D) morphants have a small clump of cardiomyocytes situated at the midline. Rtf1 morphants have no detectable cmlc2 expression at 24 hpf (E). (F–O) Dorsal views of nkx2.5 expression in 16S (F–J) and 20S (K–O) stage wild type and morphant embryos. In wild type embryos, myocardial progenitors arise in the LPM, migrate towards the midline, and fuse to form a cardiac cone (F and K). In ctr9 morphants (G and L), nkx2.5 expression is reduced and myocardial migration and fusion are delayed. Cdc73 morphants (H and M) also display delays in the migration and fusion of the myocardial progenitors, but have relatively normal nkx2.5 expression levels. Paf1 morphants (I and N) and rtf1 morphants (J and O), like ctr9 morphants, display reduced nkx2.5 expression and delayed migration and fusion of the myocardial progenitors. Expression of nkx2.5 in rtf1 morphants is barely detectable.
Reprinted from Developmental Biology, 353(1), Langenbacher, A.D., Nguyen, C.T., Cavanaugh, A.M., Huang, J., Lu, F., and Chen, J.N., The PAF1 complex differentially regulates cardiomyocyte specification, 19-28, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.