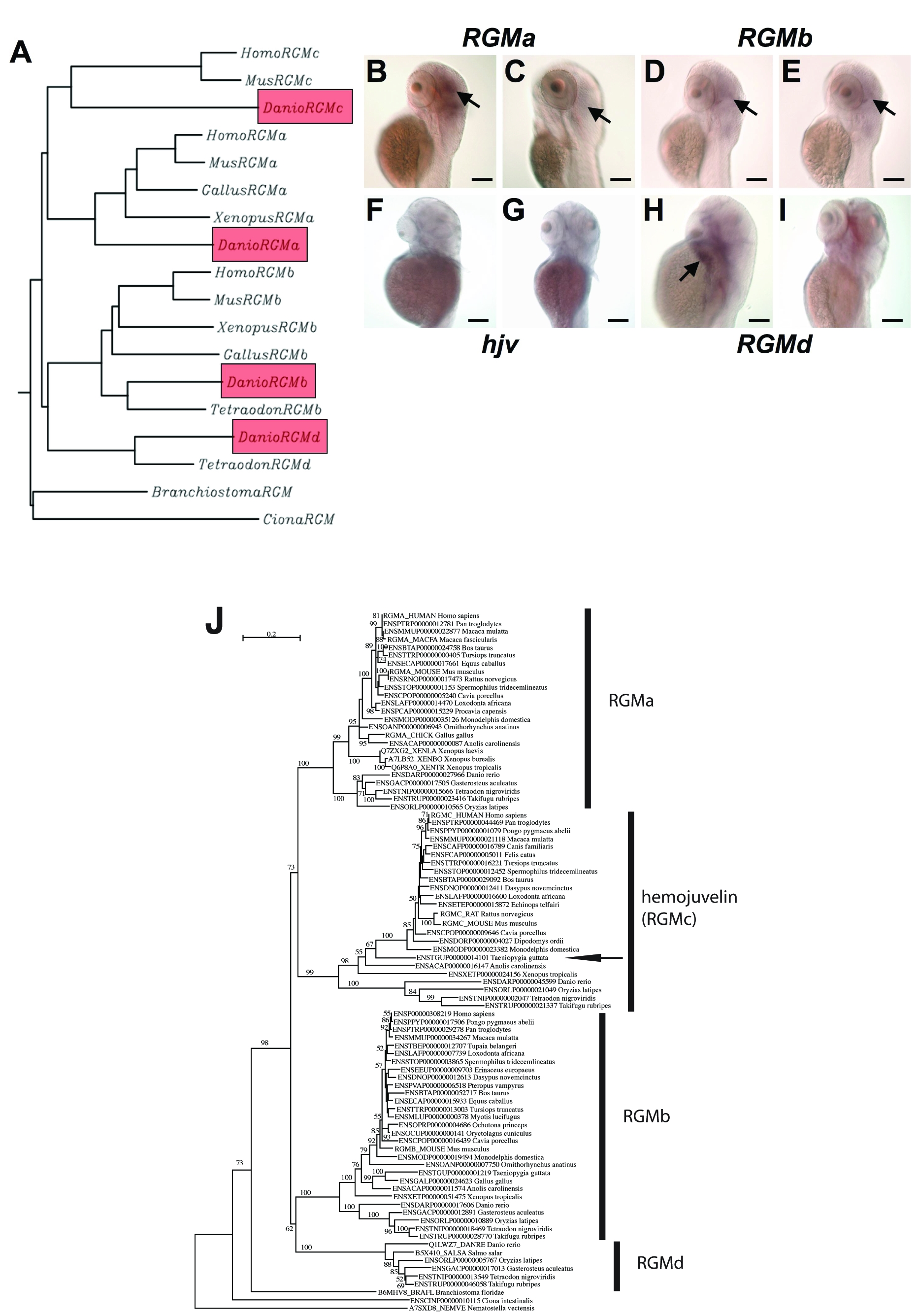
Fig. S6 Phylogeny and expression of zebrafish RGM′s. Phylogenetic tree (A) of hjv and repulsive guidance molecule genes (RGM′s) in chordates. The four zebrafish RGM paralogs are highlighted in red. Hjv is also known as RGMc. B–I. Whole mount in situ hybridization of zebrafish embryos, dorsolateral views, at 50 hpf (B,D,F,H) and 72 hpf (C,E,G,I), for RGMa (B,C), RGMb (D,E), hjv (F,G), and RGMd (H,I) revealed that none of the RGM genes are detectable in the developing liver. Strong staining was detected in the mid and hindbrain for RGMa at 50 hpf (B) and 72 hpf (C, black arrows). At 50 hpf (D) and 72 hpf (E), RGMb is faintly expressed in the mid and hindbrain (black arrows). At 50 and 72 hpf, hemojuvelin is no longer detected in the developing embryo by in situ hybridization (F,G). At 50 hpf, RGMd transcripts were detected in the pharyngeal arches (H, black arrow). RGMd expression was no longer detected at 72 hpf (I). N = 20 embryos per group. (J) Phylogenetic tree of the RGM gene family constructed with all available vertebrate sequences. Note that hjv is expressed in a wide range of mammals, fish, and in Xenopus. We have identified hjv in the genome of a bird, the zebra finch (arrow), for the first time. RGMd has only been identified in fish. To generate the tree shown, we downloaded the protein sequences of the RGM gene families defined in the Ensembl database version 52 (as of December 2008) (<http://www.ensembl.org/>), which includes the hjv sequences. In addition to the Ensembl data, which also includes the Uniprot database (<http://www.uniprot.org/>), we also screened the NCBI database (<http://www.ncbi.nlm.nih.gov/>). Alignments were generated using ClustalW and Muscle[5], [6], followed by manual refinement using SeaView[7] to remove redundant and improperly annotated sequences. Phylogenetic tree reconstruction was carried out using the maximum likelihood (ML) method. Of note, the neighbor-joining (NJ) method[7] gives the same basal node topology. For ML analyses, robustness of the obtained tree topologies was assessed with 1000 bootstrap replicates; those below 50% are not shown. The NJ tree was constructed with Phylo_Win using a Poisson correction and pairwise gap removal[7]. The ML tree was obtained with PhyML[8] using a JTT model[9], a discrete gamma model with 4 categories. The gamma shape parameter was estimated by ML and the proportion of invariable sites was also estimated by ML.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ PLoS One