Fig. 1
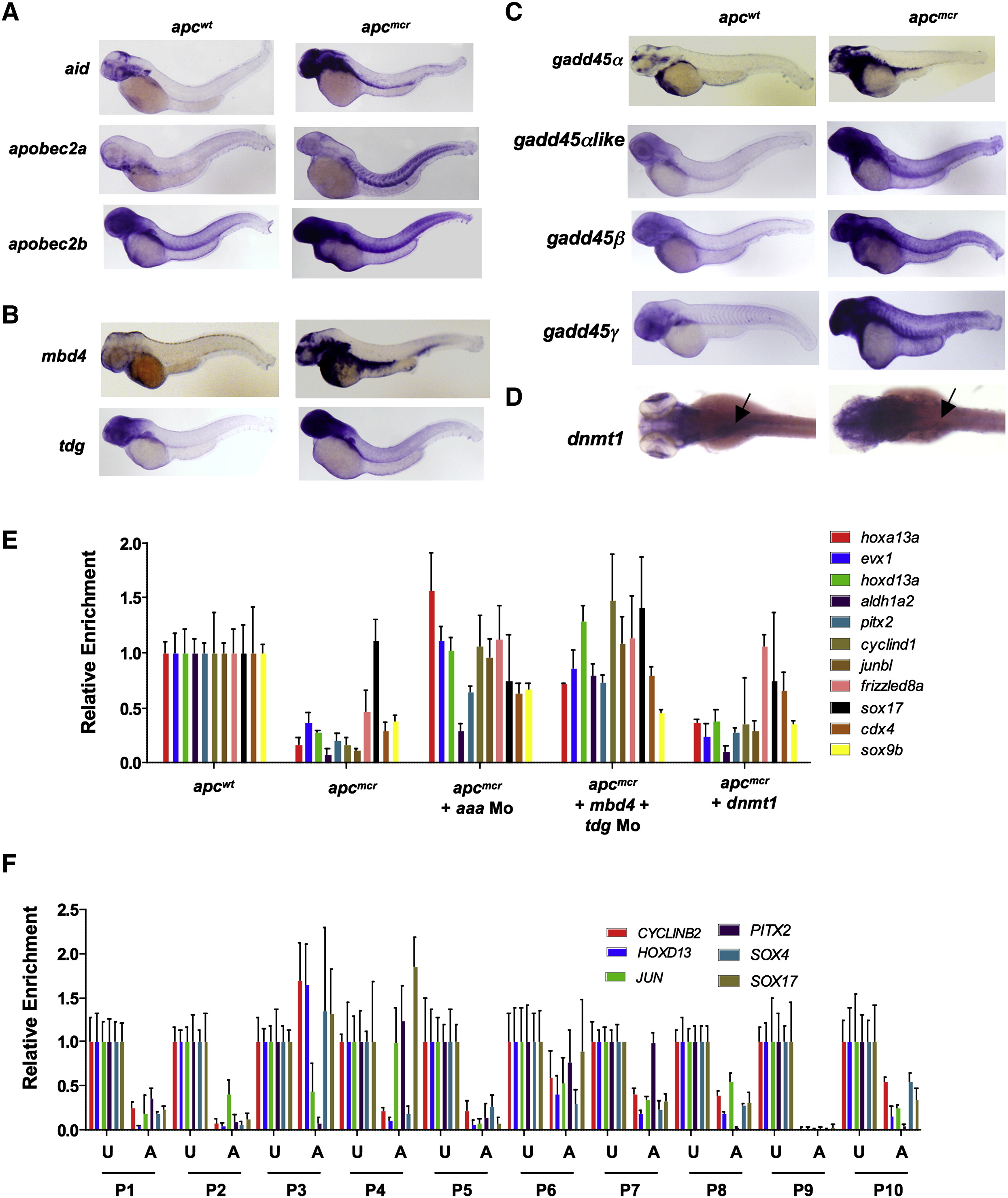
Fig. 1 Upregulation of DNA Demethylase Components, and Hypomethylated Genes in apc Mutant Embryos and FAP Adenomas
(A–D) Whole-mount in situ staining for aid (A), apobec2a (A), apobec2b (A), mbd4 (B), tdg (B), gadd45α (C), gadd45βlike (C), gadd45β (C), gadd45γ (C), and dnmt1 (D) in apc mutants (apcmcr) and siblings (apcwt) zebrafish embryos at 72 hpf.
(E) MeDIP-qPCR for genes shown (selected from genome-wide MeDIP-ChIP microarray analysis) in apcmcr and apcwt (72 hpf) that are either uninjected or injected with aaa Mo (combination of aid, apobec2a, and apobec2b morpholinos; 0.5 ng each) or mbd4 and tdg morpholinos together (1 ng each) or V5-Dnmt1 expressing plasmid (1 pg, an amount that rescues Dnmt1 morphants with 97% knockdown of Dnmt1 levels). Dnmt1 protein levels are in Figure S1C.
(F) MeDIP-qPCR for genes shown (selected from genome-wide MeDIP-ChIP microarray analysis) in human adenomas (A) and matching uninvolved tissues (U) from FAP patients. P1-P10 refers to ten different patients.
In both (E) and (F), the y axis shows values for each promoter region normalized to a negative control region lacking CpGs and then normalized to the values from wild-type or uninvolved, valued at 1.
Error bars indicate ± standard deviation (SD). See also Figure S1.
Reprinted from Cell, 142(6), Rai, K., Sarkar, S., Broadbent, T.J., Voas, M., Grossmann, K.F., Nadauld, L.D., Dehghanizadeh, S., Hagos, F.T., Li, Y., Toth, R.K., Chidester, S., Bahr, T.M., Johnson, W.E., Sklow, B., Burt, R., Cairns, B.R., and Jones, D.A., DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC, 930-942, Copyright (2010) with permission from Elsevier. Full text @ Cell