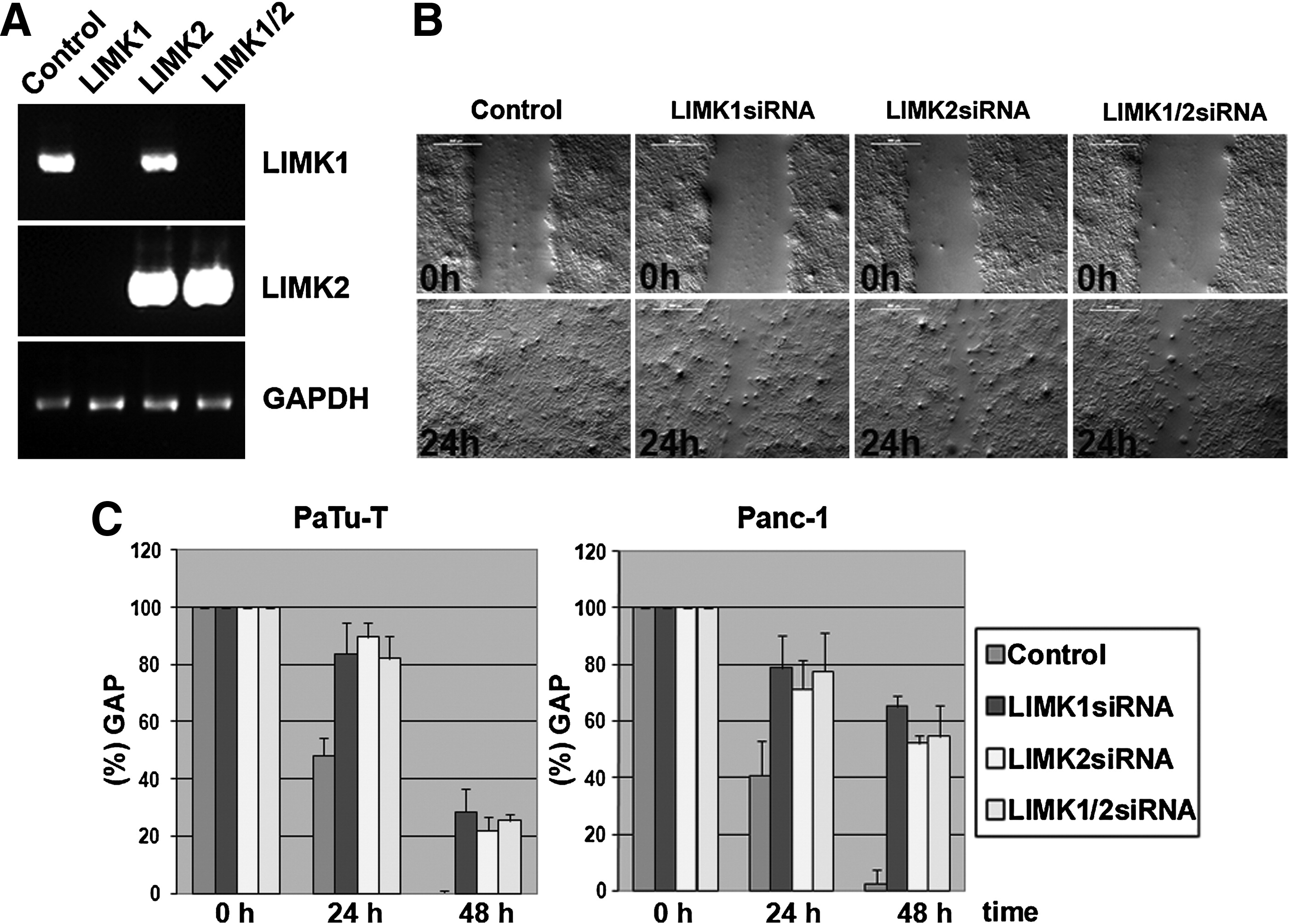
Fig. 1 Semiquantitative RT-PCR and in vitro migration assay of pancreatic cancer cell lines. (A) Semiquantitative RT-PCR shows that LIMK1 and LIMK2 are expressed in PaTu-T, and gene-specific siRNAs for LIMK1 and LIMK2 silence the respective genes by abolishing mRNA expression. Control GAPDH expression was analyzed by RT-PCR of the same RNA samples. This loading control is shown in the lower panel. LIMK1-specific primers were used in the RT-PCR shown in the top panel and LIMK2-specific amplification is shown in the middle panel. (B) An in vitro migration assay (“scratch” assay) shows differences in migration of PaTu-T cells treated with control siRNA (Control), LIMK1 siRNA, LIMK2 siRNA, or LIMK1 plus LIMK2 siRNA (LIMK1+2). (C) Gap closure (gap width) over time is shown in percentages compared with the 0h time point (set to 100%). Error bars show standard deviations between three independent experiments. On average, on day 1 (24h) the gap closure for LIMK1 was 16.4%, LIMK2 10.3%, LIMK1/2 18%, and the control 51.8%. On day 2 (48h) the gap closure for LIMK1 was 53.6%, LIMK2 10.3%, LIMK1/2 18%, and the control 51.8%. Similar results were obtained in three independent experiments and a Student′s t-test was performed, which showed that all changes observed were significant (with p-values <0.01). PCR, polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Zebrafish